| Full name: ATPase phospholipid transporting 10D (putative) | Alias Symbol: ATPVD|KIAA1487 | ||
| Type: protein-coding gene | Cytoband: 4p12 | ||
| Entrez ID: 57205 | HGNC ID: HGNC:13549 | Ensembl Gene: ENSG00000145246 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ATP10D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP10D | 57205 | 213238_at | -0.3551 | 0.7140 | |
| GSE20347 | ATP10D | 57205 | 213238_at | -0.7741 | 0.0001 | |
| GSE23400 | ATP10D | 57205 | 213238_at | -0.1158 | 0.3927 | |
| GSE26886 | ATP10D | 57205 | 213238_at | -0.1979 | 0.5262 | |
| GSE29001 | ATP10D | 57205 | 213238_at | -0.1522 | 0.7482 | |
| GSE38129 | ATP10D | 57205 | 213238_at | -0.3779 | 0.0543 | |
| GSE45670 | ATP10D | 57205 | 213238_at | 0.0455 | 0.8221 | |
| GSE53622 | ATP10D | 57205 | 161376 | -0.3614 | 0.0040 | |
| GSE53624 | ATP10D | 57205 | 161376 | -0.3980 | 0.0006 | |
| GSE63941 | ATP10D | 57205 | 213238_at | -2.1568 | 0.0059 | |
| GSE77861 | ATP10D | 57205 | 213238_at | -0.4517 | 0.1641 | |
| GSE97050 | ATP10D | 57205 | A_33_P3311473 | -0.0320 | 0.9135 | |
| SRP007169 | ATP10D | 57205 | RNAseq | -0.2857 | 0.5846 | |
| SRP008496 | ATP10D | 57205 | RNAseq | -0.3735 | 0.3019 | |
| SRP064894 | ATP10D | 57205 | RNAseq | -0.3196 | 0.2090 | |
| SRP133303 | ATP10D | 57205 | RNAseq | 0.2016 | 0.4443 | |
| SRP159526 | ATP10D | 57205 | RNAseq | -0.8083 | 0.0733 | |
| SRP193095 | ATP10D | 57205 | RNAseq | -0.3463 | 0.0920 | |
| SRP219564 | ATP10D | 57205 | RNAseq | -0.7903 | 0.1456 | |
| TCGA | ATP10D | 57205 | RNAseq | 0.0879 | 0.2529 |
Upregulated datasets: 0; Downregulated datasets: 1.
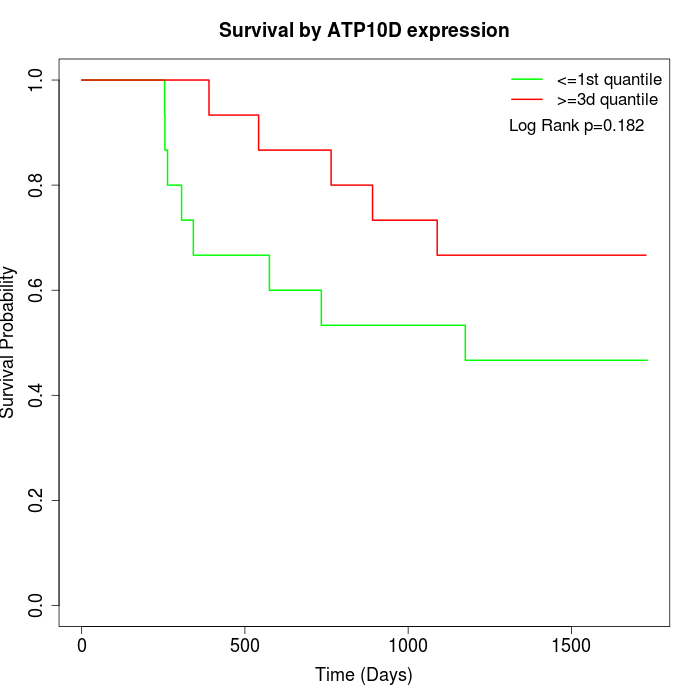
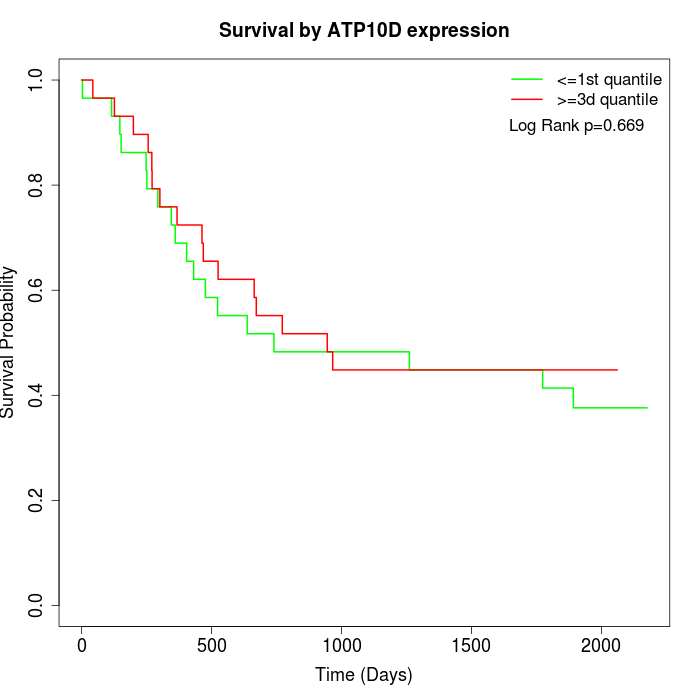
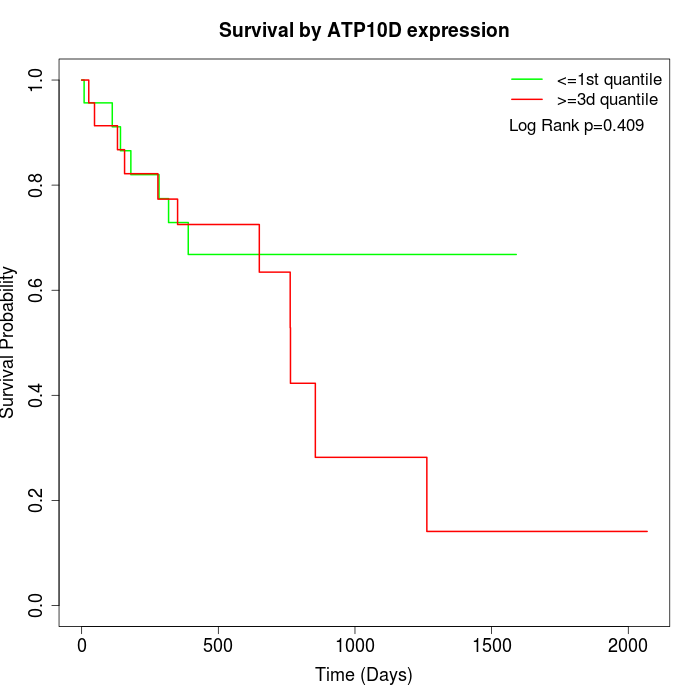
Survival by ATP10D expression:
Note: Click image to view full size file.
Copy number change of ATP10D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP10D | 57205 | 1 | 15 | 14 | |
| GSE20123 | ATP10D | 57205 | 1 | 15 | 14 | |
| GSE43470 | ATP10D | 57205 | 0 | 13 | 30 | |
| GSE46452 | ATP10D | 57205 | 1 | 37 | 21 | |
| GSE47630 | ATP10D | 57205 | 1 | 18 | 21 | |
| GSE54993 | ATP10D | 57205 | 7 | 0 | 63 | |
| GSE54994 | ATP10D | 57205 | 4 | 9 | 40 | |
| GSE60625 | ATP10D | 57205 | 0 | 0 | 11 | |
| GSE74703 | ATP10D | 57205 | 0 | 11 | 25 | |
| GSE74704 | ATP10D | 57205 | 0 | 9 | 11 | |
| TCGA | ATP10D | 57205 | 16 | 33 | 47 |
Total number of gains: 31; Total number of losses: 160; Total Number of normals: 297.
Somatic mutations of ATP10D:
Generating mutation plots.
Highly correlated genes for ATP10D:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP10D | CLTB | 0.6836 | 5 | 0 | 5 |
| ATP10D | CARHSP1 | 0.653318 | 5 | 0 | 4 |
| ATP10D | TTC37 | 0.653124 | 6 | 0 | 5 |
| ATP10D | PGM2 | 0.647899 | 5 | 0 | 4 |
| ATP10D | PLIN3 | 0.646903 | 7 | 0 | 5 |
| ATP10D | CITED2 | 0.639865 | 3 | 0 | 3 |
| ATP10D | SPDYA | 0.636326 | 3 | 0 | 3 |
| ATP10D | CDK7 | 0.634691 | 6 | 0 | 4 |
| ATP10D | DOK4 | 0.634425 | 6 | 0 | 5 |
| ATP10D | ARF4 | 0.628204 | 5 | 0 | 4 |
| ATP10D | PRRC1 | 0.628084 | 5 | 0 | 3 |
| ATP10D | INO80C | 0.627142 | 5 | 0 | 3 |
| ATP10D | TMEM161B | 0.626313 | 3 | 0 | 3 |
| ATP10D | SLC39A2 | 0.625148 | 6 | 0 | 5 |
| ATP10D | HOXA1 | 0.622893 | 5 | 0 | 4 |
| ATP10D | CEACAM1 | 0.619657 | 3 | 0 | 3 |
| ATP10D | CAPG | 0.612452 | 7 | 0 | 5 |
| ATP10D | ARSI | 0.609314 | 3 | 0 | 3 |
| ATP10D | IL18 | 0.608337 | 6 | 0 | 4 |
| ATP10D | TOR4A | 0.608107 | 4 | 0 | 3 |
For details and further investigation, click here