| Full name: clathrin light chain B | Alias Symbol: Lcb | ||
| Type: protein-coding gene | Cytoband: 5q35.2 | ||
| Entrez ID: 1212 | HGNC ID: HGNC:2091 | Ensembl Gene: ENSG00000175416 | OMIM ID: 118970 |
| Drug and gene relationship at DGIdb | |||
CLTB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05016 | Huntington's disease |
Expression of CLTB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLTB | 1212 | 211043_s_at | -1.0869 | 0.1779 | |
| GSE20347 | CLTB | 1212 | 211043_s_at | -1.5142 | 0.0000 | |
| GSE23400 | CLTB | 1212 | 211043_s_at | -0.9037 | 0.0000 | |
| GSE26886 | CLTB | 1212 | 211043_s_at | -1.4161 | 0.0000 | |
| GSE29001 | CLTB | 1212 | 211043_s_at | -1.1027 | 0.0018 | |
| GSE38129 | CLTB | 1212 | 211043_s_at | -1.0128 | 0.0000 | |
| GSE45670 | CLTB | 1212 | 211043_s_at | -0.3737 | 0.1674 | |
| GSE53622 | CLTB | 1212 | 166176 | -1.0409 | 0.0000 | |
| GSE53624 | CLTB | 1212 | 166176 | -0.8660 | 0.0000 | |
| GSE63941 | CLTB | 1212 | 211043_s_at | 0.3108 | 0.5042 | |
| GSE77861 | CLTB | 1212 | 211043_s_at | -1.3797 | 0.0155 | |
| GSE97050 | CLTB | 1212 | A_33_P3328666 | -0.1666 | 0.5806 | |
| SRP007169 | CLTB | 1212 | RNAseq | -2.7483 | 0.0000 | |
| SRP008496 | CLTB | 1212 | RNAseq | -2.9754 | 0.0000 | |
| SRP064894 | CLTB | 1212 | RNAseq | -1.3300 | 0.0000 | |
| SRP133303 | CLTB | 1212 | RNAseq | -1.0488 | 0.0005 | |
| SRP159526 | CLTB | 1212 | RNAseq | -1.6180 | 0.0017 | |
| SRP193095 | CLTB | 1212 | RNAseq | -1.7234 | 0.0000 | |
| SRP219564 | CLTB | 1212 | RNAseq | -1.3653 | 0.0038 | |
| TCGA | CLTB | 1212 | RNAseq | -0.0472 | 0.5881 |
Upregulated datasets: 0; Downregulated datasets: 13.
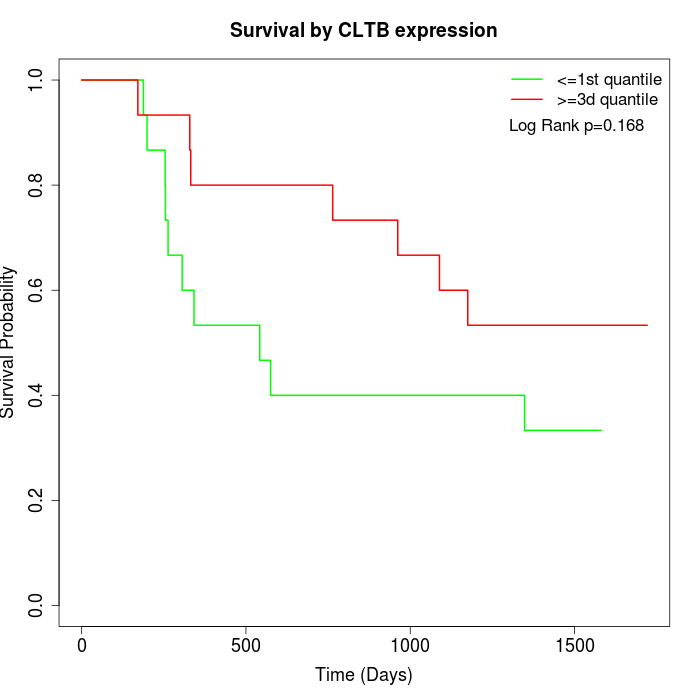
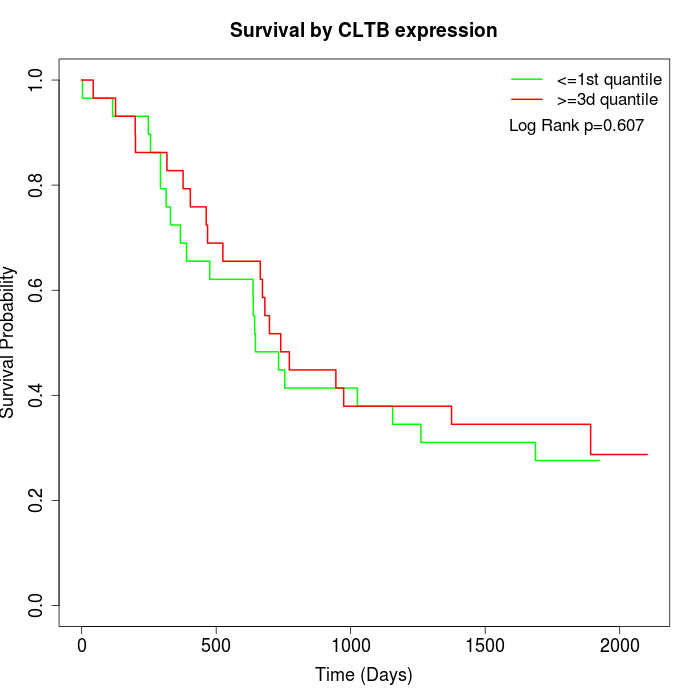
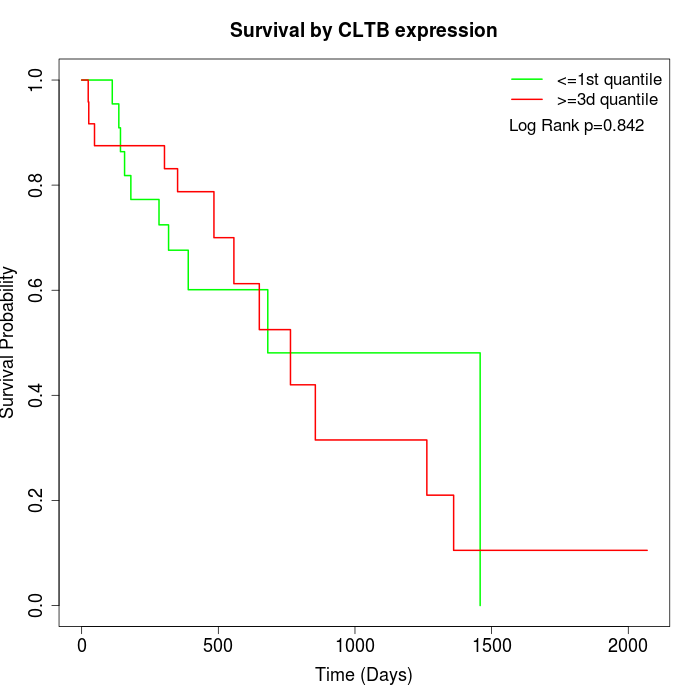
Survival by CLTB expression:
Note: Click image to view full size file.
Copy number change of CLTB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLTB | 1212 | 2 | 13 | 15 | |
| GSE20123 | CLTB | 1212 | 2 | 13 | 15 | |
| GSE43470 | CLTB | 1212 | 1 | 11 | 31 | |
| GSE46452 | CLTB | 1212 | 0 | 27 | 32 | |
| GSE47630 | CLTB | 1212 | 0 | 20 | 20 | |
| GSE54993 | CLTB | 1212 | 10 | 2 | 58 | |
| GSE54994 | CLTB | 1212 | 2 | 17 | 34 | |
| GSE60625 | CLTB | 1212 | 1 | 0 | 10 | |
| GSE74703 | CLTB | 1212 | 1 | 7 | 28 | |
| GSE74704 | CLTB | 1212 | 1 | 6 | 13 | |
| TCGA | CLTB | 1212 | 7 | 35 | 54 |
Total number of gains: 27; Total number of losses: 151; Total Number of normals: 310.
Somatic mutations of CLTB:
Generating mutation plots.
Highly correlated genes for CLTB:
Showing top 20/1652 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLTB | ALKBH7 | 0.800993 | 3 | 0 | 3 |
| CLTB | PDLIM2 | 0.797728 | 9 | 0 | 9 |
| CLTB | NLRX1 | 0.793315 | 10 | 0 | 10 |
| CLTB | LYPD3 | 0.786898 | 11 | 0 | 11 |
| CLTB | PKP3 | 0.78479 | 10 | 0 | 10 |
| CLTB | APOB | 0.77459 | 4 | 0 | 4 |
| CLTB | ACAA1 | 0.772545 | 9 | 0 | 8 |
| CLTB | ZNF185 | 0.767622 | 11 | 0 | 11 |
| CLTB | S100A14 | 0.767199 | 11 | 0 | 11 |
| CLTB | CYSRT1 | 0.766133 | 6 | 0 | 6 |
| CLTB | IL18 | 0.76576 | 10 | 0 | 10 |
| CLTB | ALS2CL | 0.761566 | 11 | 0 | 10 |
| CLTB | CWH43 | 0.761051 | 10 | 0 | 10 |
| CLTB | LRP10 | 0.75922 | 11 | 0 | 11 |
| CLTB | TMEM79 | 0.758872 | 7 | 0 | 7 |
| CLTB | APC | 0.757965 | 5 | 0 | 5 |
| CLTB | PIM1 | 0.756441 | 12 | 0 | 11 |
| CLTB | SLC25A46 | 0.75531 | 6 | 0 | 6 |
| CLTB | TOM1 | 0.754773 | 12 | 0 | 11 |
| CLTB | CCNYL1 | 0.754547 | 5 | 0 | 5 |
For details and further investigation, click here