| Full name: tetratricopeptide repeat domain 37 | Alias Symbol: THES | ||
| Type: protein-coding gene | Cytoband: 5q15 | ||
| Entrez ID: 9652 | HGNC ID: HGNC:23639 | Ensembl Gene: ENSG00000198677 | OMIM ID: 614589 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TTC37:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TTC37 | 9652 | 203049_s_at | -0.5582 | 0.2312 | |
| GSE20347 | TTC37 | 9652 | 203049_s_at | -0.6797 | 0.0001 | |
| GSE23400 | TTC37 | 9652 | 203048_s_at | -0.0847 | 0.4315 | |
| GSE26886 | TTC37 | 9652 | 203049_s_at | -0.1580 | 0.5987 | |
| GSE29001 | TTC37 | 9652 | 203049_s_at | -0.1766 | 0.7524 | |
| GSE38129 | TTC37 | 9652 | 203049_s_at | -0.6282 | 0.0000 | |
| GSE45670 | TTC37 | 9652 | 203049_s_at | -0.2425 | 0.1157 | |
| GSE53622 | TTC37 | 9652 | 47871 | -0.4684 | 0.0000 | |
| GSE53624 | TTC37 | 9652 | 47871 | -0.2300 | 0.0000 | |
| GSE63941 | TTC37 | 9652 | 203049_s_at | -1.2423 | 0.0151 | |
| GSE77861 | TTC37 | 9652 | 203049_s_at | -1.0433 | 0.0027 | |
| GSE97050 | TTC37 | 9652 | A_33_P3259620 | -0.3469 | 0.2434 | |
| SRP007169 | TTC37 | 9652 | RNAseq | -0.9693 | 0.0031 | |
| SRP008496 | TTC37 | 9652 | RNAseq | -0.8126 | 0.0000 | |
| SRP064894 | TTC37 | 9652 | RNAseq | -0.2416 | 0.3448 | |
| SRP133303 | TTC37 | 9652 | RNAseq | -0.0536 | 0.7961 | |
| SRP159526 | TTC37 | 9652 | RNAseq | -0.6825 | 0.0003 | |
| SRP193095 | TTC37 | 9652 | RNAseq | -0.6892 | 0.0000 | |
| SRP219564 | TTC37 | 9652 | RNAseq | -0.4781 | 0.1610 | |
| TCGA | TTC37 | 9652 | RNAseq | -0.2449 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 2.
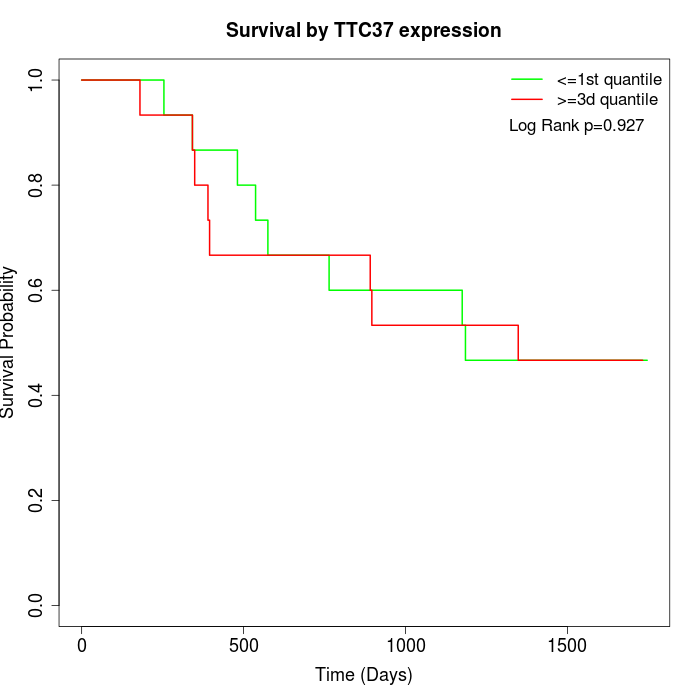
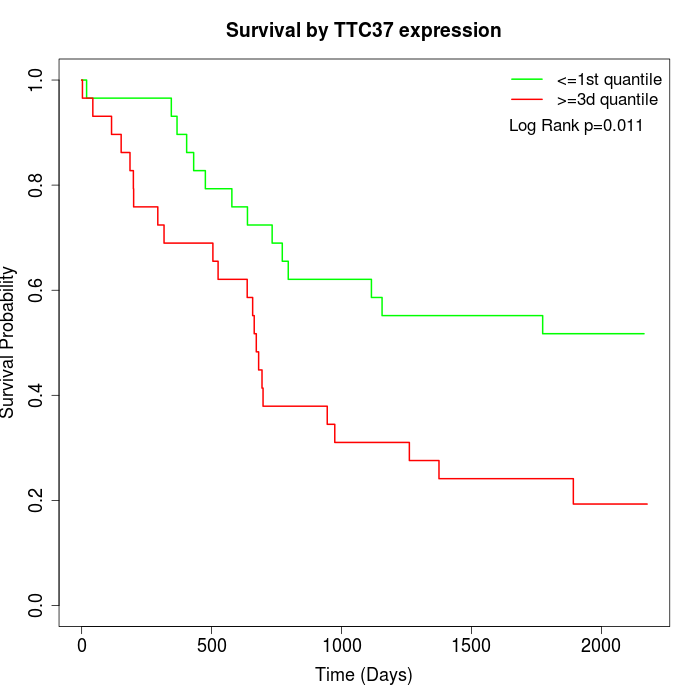
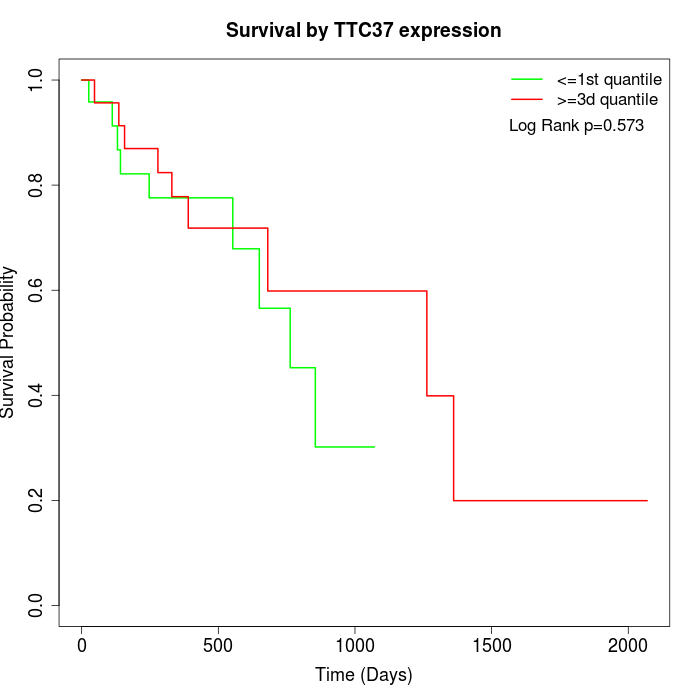
Survival by TTC37 expression:
Note: Click image to view full size file.
Copy number change of TTC37:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TTC37 | 9652 | 1 | 13 | 16 | |
| GSE20123 | TTC37 | 9652 | 1 | 13 | 16 | |
| GSE43470 | TTC37 | 9652 | 3 | 9 | 31 | |
| GSE46452 | TTC37 | 9652 | 0 | 28 | 31 | |
| GSE47630 | TTC37 | 9652 | 0 | 20 | 20 | |
| GSE54993 | TTC37 | 9652 | 8 | 1 | 61 | |
| GSE54994 | TTC37 | 9652 | 1 | 18 | 34 | |
| GSE60625 | TTC37 | 9652 | 0 | 0 | 11 | |
| GSE74703 | TTC37 | 9652 | 2 | 7 | 27 | |
| GSE74704 | TTC37 | 9652 | 1 | 7 | 12 | |
| TCGA | TTC37 | 9652 | 3 | 45 | 48 |
Total number of gains: 20; Total number of losses: 161; Total Number of normals: 307.
Somatic mutations of TTC37:
Generating mutation plots.
Highly correlated genes for TTC37:
Showing top 20/1089 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TTC37 | ACBD5 | 0.756479 | 3 | 0 | 3 |
| TTC37 | NUP54 | 0.754276 | 3 | 0 | 3 |
| TTC37 | METAP2 | 0.74826 | 3 | 0 | 3 |
| TTC37 | ANKRD37 | 0.743574 | 3 | 0 | 3 |
| TTC37 | CAP1 | 0.739336 | 3 | 0 | 3 |
| TTC37 | MRPL40 | 0.732992 | 3 | 0 | 3 |
| TTC37 | PNPLA8 | 0.731417 | 3 | 0 | 3 |
| TTC37 | EIF3I | 0.730106 | 3 | 0 | 3 |
| TTC37 | ZMAT3 | 0.727815 | 3 | 0 | 3 |
| TTC37 | RNF139 | 0.727304 | 3 | 0 | 3 |
| TTC37 | HPS5 | 0.726193 | 3 | 0 | 3 |
| TTC37 | BHLHE40 | 0.725242 | 4 | 0 | 3 |
| TTC37 | RAP1GDS1 | 0.71813 | 5 | 0 | 4 |
| TTC37 | CNST | 0.71743 | 3 | 0 | 3 |
| TTC37 | AHNAK2 | 0.716866 | 3 | 0 | 3 |
| TTC37 | BVES | 0.716621 | 3 | 0 | 3 |
| TTC37 | SREK1IP1 | 0.716185 | 8 | 0 | 6 |
| TTC37 | CWH43 | 0.715974 | 4 | 0 | 4 |
| TTC37 | C6orf120 | 0.715623 | 4 | 0 | 4 |
| TTC37 | SNX18 | 0.715414 | 3 | 0 | 3 |
For details and further investigation, click here