| Full name: ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2 | Alias Symbol: SERCA2 | ||
| Type: protein-coding gene | Cytoband: 12q24.11 | ||
| Entrez ID: 488 | HGNC ID: HGNC:812 | Ensembl Gene: ENSG00000174437 | OMIM ID: 108740 |
| Drug and gene relationship at DGIdb | |||
ATP2A2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa04919 | Thyroid hormone signaling pathway |
Expression of ATP2A2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP2A2 | 488 | 209186_at | 0.6853 | 0.1009 | |
| GSE20347 | ATP2A2 | 488 | 209186_at | 0.1976 | 0.2149 | |
| GSE23400 | ATP2A2 | 488 | 209186_at | 0.3596 | 0.0000 | |
| GSE26886 | ATP2A2 | 488 | 212361_s_at | -0.3795 | 0.2833 | |
| GSE29001 | ATP2A2 | 488 | 209186_at | -0.0292 | 0.8973 | |
| GSE38129 | ATP2A2 | 488 | 209186_at | 0.6178 | 0.0075 | |
| GSE45670 | ATP2A2 | 488 | 209186_at | 0.5700 | 0.0000 | |
| GSE53622 | ATP2A2 | 488 | 63672 | 0.4021 | 0.0000 | |
| GSE53624 | ATP2A2 | 488 | 63672 | 0.2756 | 0.0000 | |
| GSE63941 | ATP2A2 | 488 | 209186_at | 1.1514 | 0.0003 | |
| GSE77861 | ATP2A2 | 488 | 209186_at | 0.2385 | 0.3049 | |
| GSE97050 | ATP2A2 | 488 | A_24_P141786 | 0.3660 | 0.1479 | |
| SRP007169 | ATP2A2 | 488 | RNAseq | 0.3200 | 0.3333 | |
| SRP008496 | ATP2A2 | 488 | RNAseq | -0.0107 | 0.9581 | |
| SRP064894 | ATP2A2 | 488 | RNAseq | -0.3186 | 0.2405 | |
| SRP133303 | ATP2A2 | 488 | RNAseq | 0.2534 | 0.0702 | |
| SRP159526 | ATP2A2 | 488 | RNAseq | 0.1971 | 0.5835 | |
| SRP193095 | ATP2A2 | 488 | RNAseq | 0.2743 | 0.0129 | |
| SRP219564 | ATP2A2 | 488 | RNAseq | -0.2955 | 0.4500 | |
| TCGA | ATP2A2 | 488 | RNAseq | 0.1135 | 0.0023 |
Upregulated datasets: 1; Downregulated datasets: 0.
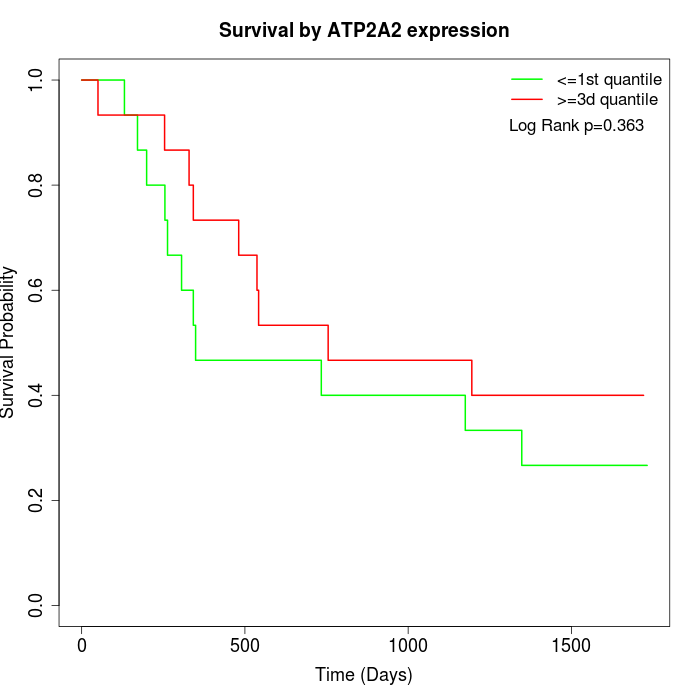
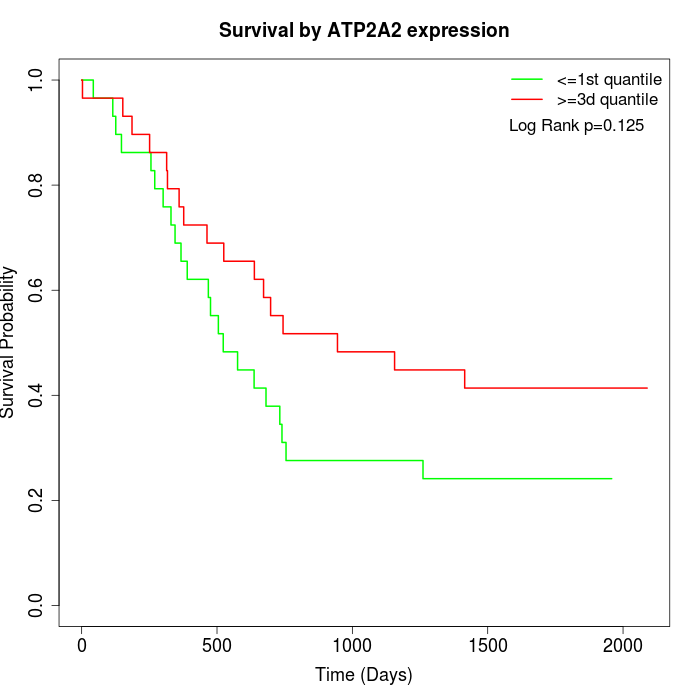
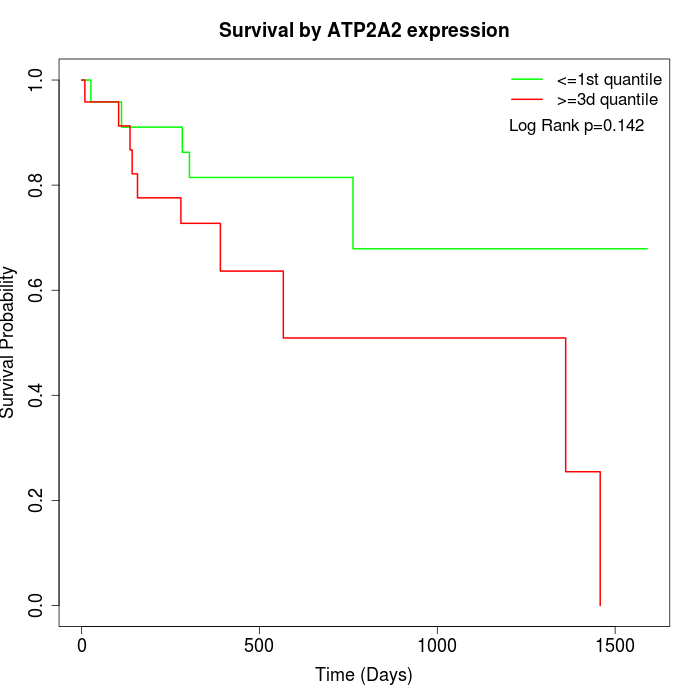
Survival by ATP2A2 expression:
Note: Click image to view full size file.
Copy number change of ATP2A2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP2A2 | 488 | 5 | 3 | 22 | |
| GSE20123 | ATP2A2 | 488 | 5 | 3 | 22 | |
| GSE43470 | ATP2A2 | 488 | 2 | 1 | 40 | |
| GSE46452 | ATP2A2 | 488 | 9 | 1 | 49 | |
| GSE47630 | ATP2A2 | 488 | 9 | 3 | 28 | |
| GSE54993 | ATP2A2 | 488 | 0 | 5 | 65 | |
| GSE54994 | ATP2A2 | 488 | 4 | 3 | 46 | |
| GSE60625 | ATP2A2 | 488 | 0 | 0 | 11 | |
| GSE74703 | ATP2A2 | 488 | 2 | 0 | 34 | |
| GSE74704 | ATP2A2 | 488 | 2 | 2 | 16 | |
| TCGA | ATP2A2 | 488 | 22 | 10 | 64 |
Total number of gains: 60; Total number of losses: 31; Total Number of normals: 397.
Somatic mutations of ATP2A2:
Generating mutation plots.
Highly correlated genes for ATP2A2:
Showing top 20/442 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP2A2 | KDF1 | 0.702235 | 4 | 0 | 4 |
| ATP2A2 | CAPN15 | 0.699508 | 4 | 0 | 4 |
| ATP2A2 | RCC2 | 0.697025 | 6 | 0 | 6 |
| ATP2A2 | CMTM4 | 0.692003 | 3 | 0 | 3 |
| ATP2A2 | NAA50 | 0.68444 | 6 | 0 | 6 |
| ATP2A2 | ADAM15 | 0.675516 | 3 | 0 | 3 |
| ATP2A2 | BTF3L4 | 0.674865 | 4 | 0 | 4 |
| ATP2A2 | LINC01000 | 0.672273 | 3 | 0 | 3 |
| ATP2A2 | MARVELD2 | 0.668957 | 3 | 0 | 3 |
| ATP2A2 | DGAT1 | 0.668931 | 3 | 0 | 3 |
| ATP2A2 | FUOM | 0.668305 | 3 | 0 | 3 |
| ATP2A2 | STXBP2 | 0.666848 | 5 | 0 | 4 |
| ATP2A2 | CNOT3 | 0.661909 | 4 | 0 | 4 |
| ATP2A2 | PPP1R35 | 0.659812 | 3 | 0 | 3 |
| ATP2A2 | TRIM29 | 0.658996 | 3 | 0 | 3 |
| ATP2A2 | TADA2A | 0.650192 | 3 | 0 | 3 |
| ATP2A2 | WDR18 | 0.647873 | 7 | 0 | 6 |
| ATP2A2 | EIF4G1 | 0.64486 | 8 | 0 | 6 |
| ATP2A2 | CCDC137 | 0.641754 | 4 | 0 | 3 |
| ATP2A2 | CDK16 | 0.639561 | 5 | 0 | 3 |
For details and further investigation, click here