| Full name: ATP synthase mitochondrial F1 complex assembly factor 2 | Alias Symbol: Atp12p|ATP12|LP3663|MGC29736 | ||
| Type: protein-coding gene | Cytoband: 17p11.2 | ||
| Entrez ID: 91647 | HGNC ID: HGNC:18802 | Ensembl Gene: ENSG00000171953 | OMIM ID: 608918 |
| Drug and gene relationship at DGIdb | |||
Expression of ATPAF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATPAF2 | 91647 | 213057_at | -0.2245 | 0.4860 | |
| GSE20347 | ATPAF2 | 91647 | 213057_at | -0.3307 | 0.0103 | |
| GSE23400 | ATPAF2 | 91647 | 213057_at | -0.1335 | 0.0022 | |
| GSE26886 | ATPAF2 | 91647 | 213057_at | 0.2333 | 0.2163 | |
| GSE29001 | ATPAF2 | 91647 | 214330_at | -0.0884 | 0.5878 | |
| GSE38129 | ATPAF2 | 91647 | 213057_at | -0.1547 | 0.1548 | |
| GSE45670 | ATPAF2 | 91647 | 213057_at | -0.0426 | 0.7995 | |
| GSE53622 | ATPAF2 | 91647 | 64265 | -0.5236 | 0.0000 | |
| GSE53624 | ATPAF2 | 91647 | 64265 | -0.3894 | 0.0000 | |
| GSE63941 | ATPAF2 | 91647 | 213057_at | 1.1347 | 0.0728 | |
| GSE77861 | ATPAF2 | 91647 | 213057_at | -0.0447 | 0.7982 | |
| GSE97050 | ATPAF2 | 91647 | A_23_P418493 | -0.1321 | 0.6332 | |
| SRP007169 | ATPAF2 | 91647 | RNAseq | -0.1661 | 0.7513 | |
| SRP008496 | ATPAF2 | 91647 | RNAseq | -0.1842 | 0.5630 | |
| SRP064894 | ATPAF2 | 91647 | RNAseq | 0.3411 | 0.1136 | |
| SRP133303 | ATPAF2 | 91647 | RNAseq | -0.0915 | 0.5038 | |
| SRP159526 | ATPAF2 | 91647 | RNAseq | 0.0389 | 0.7983 | |
| SRP193095 | ATPAF2 | 91647 | RNAseq | -0.3258 | 0.0003 | |
| SRP219564 | ATPAF2 | 91647 | RNAseq | 0.2786 | 0.3948 | |
| TCGA | ATPAF2 | 91647 | RNAseq | -0.1162 | 0.0637 |
Upregulated datasets: 0; Downregulated datasets: 0.
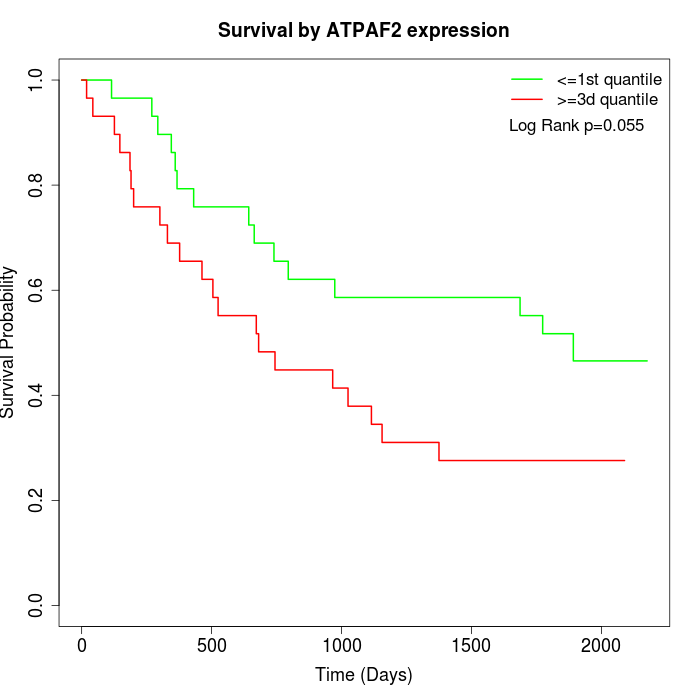
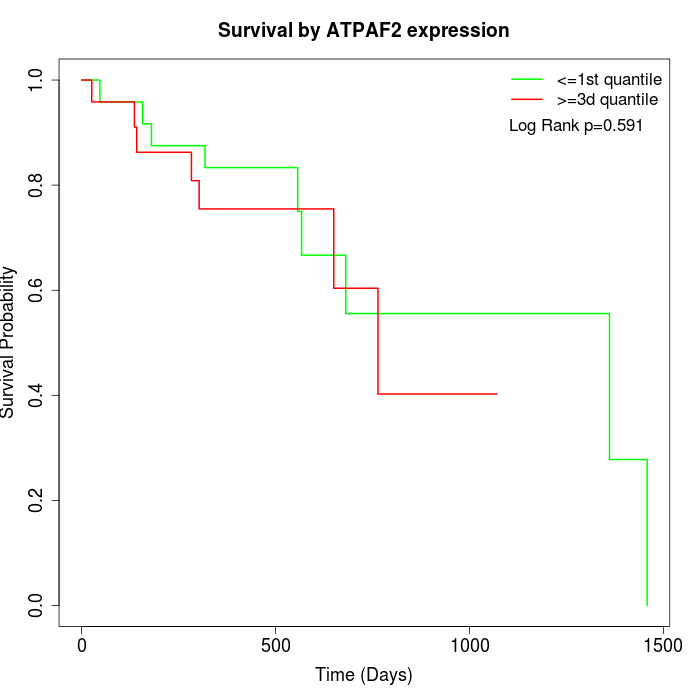
Survival by ATPAF2 expression:
Note: Click image to view full size file.
Copy number change of ATPAF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATPAF2 | 91647 | 3 | 3 | 24 | |
| GSE20123 | ATPAF2 | 91647 | 3 | 4 | 23 | |
| GSE43470 | ATPAF2 | 91647 | 1 | 5 | 37 | |
| GSE46452 | ATPAF2 | 91647 | 34 | 1 | 24 | |
| GSE47630 | ATPAF2 | 91647 | 7 | 1 | 32 | |
| GSE54993 | ATPAF2 | 91647 | 3 | 3 | 64 | |
| GSE54994 | ATPAF2 | 91647 | 6 | 6 | 41 | |
| GSE60625 | ATPAF2 | 91647 | 4 | 0 | 7 | |
| GSE74703 | ATPAF2 | 91647 | 1 | 2 | 33 | |
| GSE74704 | ATPAF2 | 91647 | 2 | 1 | 17 | |
| TCGA | ATPAF2 | 91647 | 18 | 23 | 55 |
Total number of gains: 82; Total number of losses: 49; Total Number of normals: 357.
Somatic mutations of ATPAF2:
Generating mutation plots.
Highly correlated genes for ATPAF2:
Showing top 20/128 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATPAF2 | ONECUT1 | 0.675443 | 3 | 0 | 3 |
| ATPAF2 | LRRC46 | 0.663384 | 3 | 0 | 3 |
| ATPAF2 | VPS25 | 0.652364 | 3 | 0 | 3 |
| ATPAF2 | CAAP1 | 0.647041 | 3 | 0 | 3 |
| ATPAF2 | CNGB1 | 0.642234 | 3 | 0 | 3 |
| ATPAF2 | SMIM15 | 0.640421 | 3 | 0 | 3 |
| ATPAF2 | SCNN1G | 0.634063 | 3 | 0 | 3 |
| ATPAF2 | C6orf132 | 0.630782 | 3 | 0 | 3 |
| ATPAF2 | TAS2R3 | 0.621794 | 3 | 0 | 3 |
| ATPAF2 | SYCE3 | 0.614032 | 3 | 0 | 3 |
| ATPAF2 | AAMP | 0.608494 | 4 | 0 | 3 |
| ATPAF2 | TECTA | 0.607521 | 4 | 0 | 3 |
| ATPAF2 | ECHDC3 | 0.597593 | 4 | 0 | 3 |
| ATPAF2 | SLC10A1 | 0.595266 | 4 | 0 | 3 |
| ATPAF2 | ACSBG2 | 0.594043 | 3 | 0 | 3 |
| ATPAF2 | EVPLL | 0.591093 | 4 | 0 | 3 |
| ATPAF2 | DNASE1 | 0.589529 | 4 | 0 | 3 |
| ATPAF2 | MIEF2 | 0.588064 | 4 | 0 | 3 |
| ATPAF2 | INO80C | 0.587799 | 4 | 0 | 3 |
| ATPAF2 | ANKRD24 | 0.58712 | 3 | 0 | 3 |
For details and further investigation, click here