| Full name: BCL2 apoptosis regulator | Alias Symbol: Bcl-2|PPP1R50 | ||
| Type: protein-coding gene | Cytoband: 18q21.33 | ||
| Entrez ID: 596 | HGNC ID: HGNC:990 | Ensembl Gene: ENSG00000171791 | OMIM ID: 151430 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BCL2 involved pathways:
Expression of BCL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCL2 | 596 | 203685_at | -0.7412 | 0.5623 | |
| GSE20347 | BCL2 | 596 | 203685_at | 0.3057 | 0.4137 | |
| GSE23400 | BCL2 | 596 | 203685_at | -0.0037 | 0.9738 | |
| GSE26886 | BCL2 | 596 | 203685_at | 0.5364 | 0.4283 | |
| GSE29001 | BCL2 | 596 | 203685_at | -0.1271 | 0.8525 | |
| GSE38129 | BCL2 | 596 | 203685_at | -0.2325 | 0.5808 | |
| GSE45670 | BCL2 | 596 | 207004_at | -0.0830 | 0.4136 | |
| GSE53622 | BCL2 | 596 | 16891 | -0.3421 | 0.0058 | |
| GSE53624 | BCL2 | 596 | 16891 | -0.2145 | 0.0082 | |
| GSE63941 | BCL2 | 596 | 207004_at | -0.2584 | 0.1748 | |
| GSE77861 | BCL2 | 596 | 207004_at | -0.0811 | 0.6727 | |
| GSE97050 | BCL2 | 596 | A_33_P3355185 | -0.3000 | 0.2502 | |
| SRP007169 | BCL2 | 596 | RNAseq | -0.0636 | 0.9271 | |
| SRP008496 | BCL2 | 596 | RNAseq | -0.4413 | 0.2663 | |
| SRP064894 | BCL2 | 596 | RNAseq | -0.9160 | 0.0016 | |
| SRP133303 | BCL2 | 596 | RNAseq | -0.5014 | 0.0443 | |
| SRP159526 | BCL2 | 596 | RNAseq | 0.1598 | 0.6747 | |
| SRP193095 | BCL2 | 596 | RNAseq | -0.3518 | 0.1253 | |
| SRP219564 | BCL2 | 596 | RNAseq | -0.1613 | 0.7826 | |
| TCGA | BCL2 | 596 | RNAseq | -0.3309 | 0.0214 |
Upregulated datasets: 0; Downregulated datasets: 0.
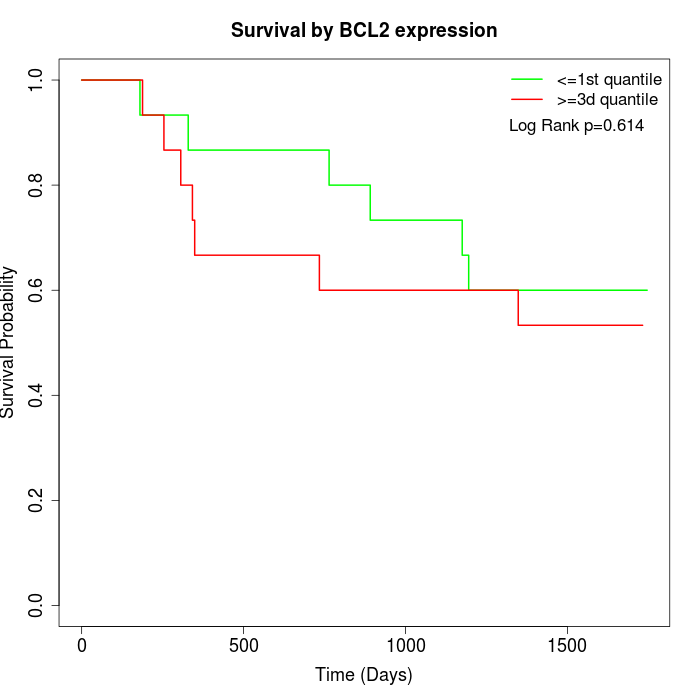
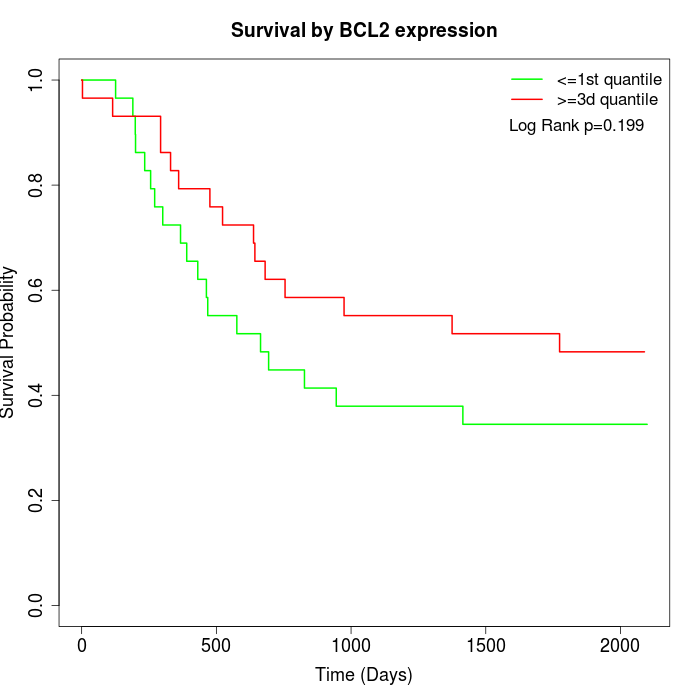
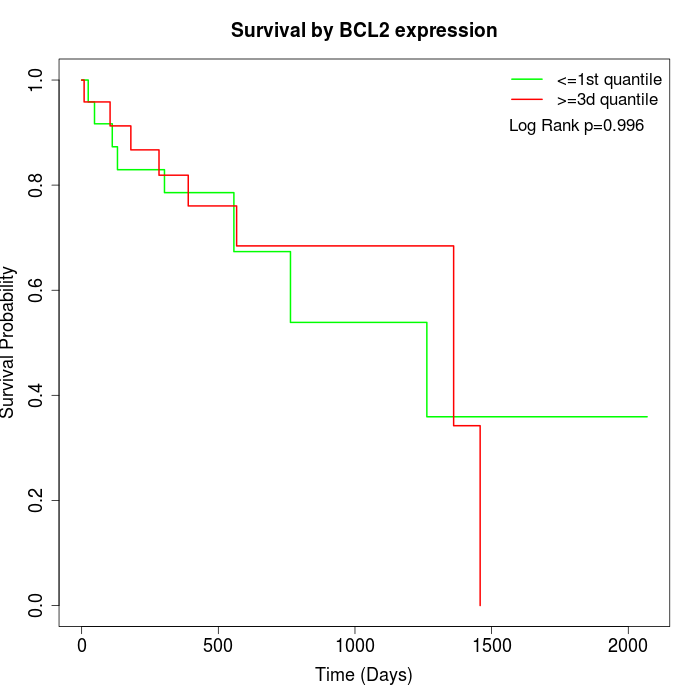
Survival by BCL2 expression:
Note: Click image to view full size file.
Copy number change of BCL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCL2 | 596 | 1 | 9 | 20 | |
| GSE20123 | BCL2 | 596 | 1 | 9 | 20 | |
| GSE43470 | BCL2 | 596 | 0 | 7 | 36 | |
| GSE46452 | BCL2 | 596 | 1 | 26 | 32 | |
| GSE47630 | BCL2 | 596 | 5 | 21 | 14 | |
| GSE54993 | BCL2 | 596 | 9 | 0 | 61 | |
| GSE54994 | BCL2 | 596 | 1 | 18 | 34 | |
| GSE60625 | BCL2 | 596 | 0 | 4 | 7 | |
| GSE74703 | BCL2 | 596 | 0 | 7 | 29 | |
| GSE74704 | BCL2 | 596 | 1 | 6 | 13 | |
| TCGA | BCL2 | 596 | 10 | 45 | 41 |
Total number of gains: 29; Total number of losses: 152; Total Number of normals: 307.
Somatic mutations of BCL2:
Generating mutation plots.
Highly correlated genes for BCL2:
Showing top 20/329 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCL2 | NUDT16 | 0.763166 | 3 | 0 | 3 |
| BCL2 | BCAT2 | 0.709991 | 3 | 0 | 3 |
| BCL2 | DISP1 | 0.709424 | 3 | 0 | 3 |
| BCL2 | GPR26 | 0.701532 | 3 | 0 | 3 |
| BCL2 | CDO1 | 0.701341 | 4 | 0 | 3 |
| BCL2 | PIP5K1B | 0.697245 | 5 | 0 | 5 |
| BCL2 | TMEM8B | 0.69561 | 6 | 0 | 6 |
| BCL2 | FGF13 | 0.692404 | 4 | 0 | 4 |
| BCL2 | ZNF358 | 0.688295 | 4 | 0 | 4 |
| BCL2 | BEND5 | 0.679418 | 7 | 0 | 6 |
| BCL2 | STXBP6 | 0.674289 | 6 | 0 | 6 |
| BCL2 | PTGIS | 0.673141 | 6 | 0 | 5 |
| BCL2 | ADCY5 | 0.671641 | 5 | 0 | 4 |
| BCL2 | FAM117A | 0.669408 | 8 | 0 | 8 |
| BCL2 | KANK2 | 0.665853 | 4 | 0 | 3 |
| BCL2 | LRRC4B | 0.663355 | 3 | 0 | 3 |
| BCL2 | MYEF2 | 0.662959 | 3 | 0 | 3 |
| BCL2 | BEX1 | 0.659409 | 5 | 0 | 4 |
| BCL2 | PROM1 | 0.655379 | 5 | 0 | 4 |
| BCL2 | TSPYL5 | 0.654015 | 5 | 0 | 3 |
For details and further investigation, click here