| Full name: fibroblast growth factor 13 | Alias Symbol: FHF2|FGF2 | ||
| Type: protein-coding gene | Cytoband: Xq26.3-q27.1 | ||
| Entrez ID: 2258 | HGNC ID: HGNC:3670 | Ensembl Gene: ENSG00000129682 | OMIM ID: 300070 |
| Drug and gene relationship at DGIdb | |||
FGF13 involved pathways:
Expression of FGF13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FGF13 | 2258 | 205110_s_at | -1.4720 | 0.3328 | |
| GSE20347 | FGF13 | 2258 | 205110_s_at | 0.9978 | 0.0624 | |
| GSE23400 | FGF13 | 2258 | 205110_s_at | -0.0214 | 0.8976 | |
| GSE26886 | FGF13 | 2258 | 205110_s_at | 0.9565 | 0.0651 | |
| GSE29001 | FGF13 | 2258 | 205110_s_at | 0.0325 | 0.9545 | |
| GSE38129 | FGF13 | 2258 | 205110_s_at | 0.1668 | 0.7717 | |
| GSE45670 | FGF13 | 2258 | 205110_s_at | -1.4495 | 0.0435 | |
| GSE53622 | FGF13 | 2258 | 90164 | -0.3403 | 0.0552 | |
| GSE53624 | FGF13 | 2258 | 35436 | -0.1679 | 0.3438 | |
| GSE63941 | FGF13 | 2258 | 205110_s_at | -3.4344 | 0.0084 | |
| GSE77861 | FGF13 | 2258 | 205110_s_at | 0.5936 | 0.0379 | |
| GSE97050 | FGF13 | 2258 | A_23_P217319 | -0.4309 | 0.4789 | |
| SRP007169 | FGF13 | 2258 | RNAseq | 1.9149 | 0.0007 | |
| SRP064894 | FGF13 | 2258 | RNAseq | -0.3178 | 0.0551 | |
| SRP133303 | FGF13 | 2258 | RNAseq | 0.2053 | 0.0470 | |
| SRP159526 | FGF13 | 2258 | RNAseq | 0.1815 | 0.5472 | |
| SRP193095 | FGF13 | 2258 | RNAseq | 0.3604 | 0.0068 | |
| SRP219564 | FGF13 | 2258 | RNAseq | -0.1846 | 0.6622 | |
| TCGA | FGF13 | 2258 | RNAseq | -0.6994 | 0.0183 |
Upregulated datasets: 1; Downregulated datasets: 2.
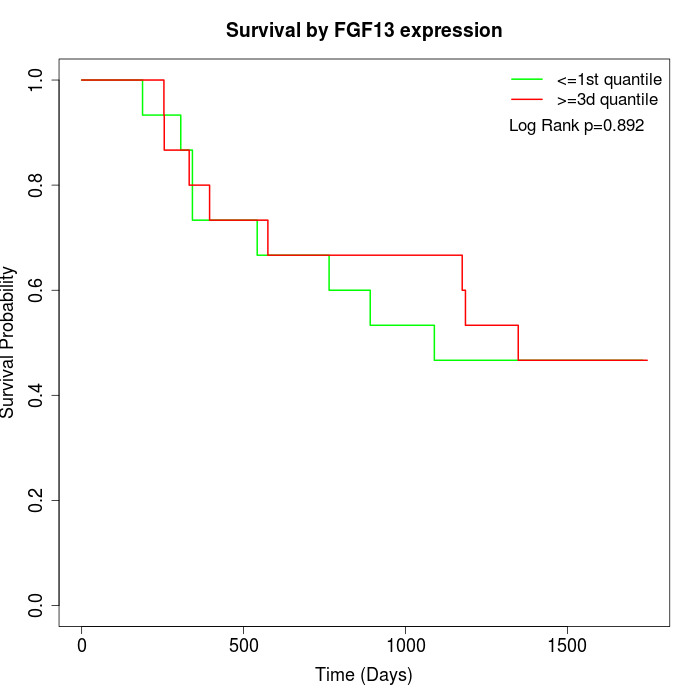
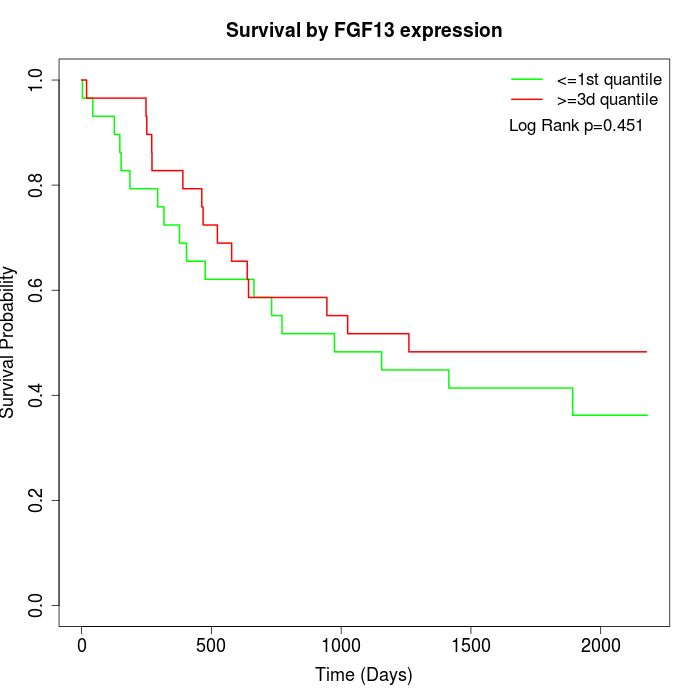
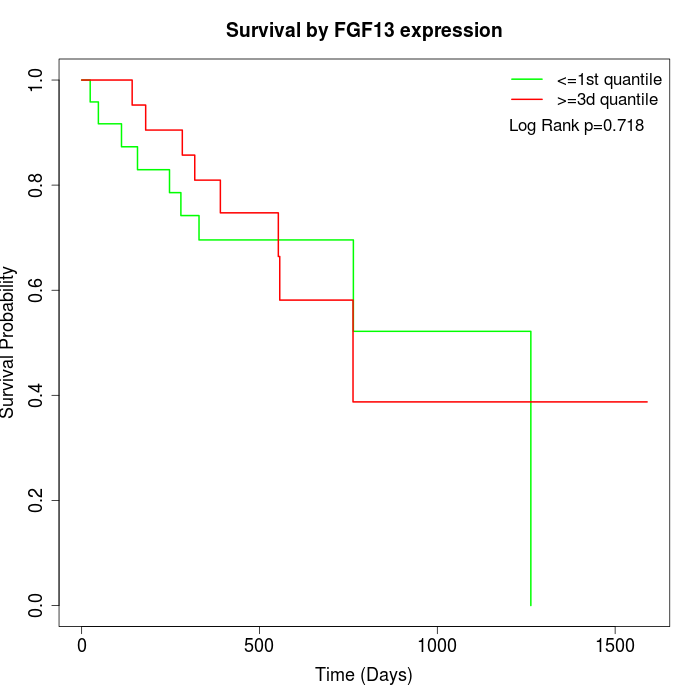
Survival by FGF13 expression:
Note: Click image to view full size file.
Copy number change of FGF13:
No record found for this gene.
Somatic mutations of FGF13:
Generating mutation plots.
Highly correlated genes for FGF13:
Showing top 20/357 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FGF13 | HECTD2 | 0.710128 | 3 | 0 | 3 |
| FGF13 | IL17D | 0.698614 | 5 | 0 | 5 |
| FGF13 | DACT3 | 0.698226 | 4 | 0 | 3 |
| FGF13 | BCL2 | 0.692404 | 4 | 0 | 4 |
| FGF13 | FAT4 | 0.691829 | 6 | 0 | 6 |
| FGF13 | GPM6A | 0.685649 | 4 | 0 | 4 |
| FGF13 | ALDH1B1 | 0.655342 | 4 | 0 | 3 |
| FGF13 | ITGB1BP2 | 0.651591 | 5 | 0 | 4 |
| FGF13 | RYR2 | 0.646786 | 6 | 0 | 5 |
| FGF13 | NRK | 0.64412 | 5 | 0 | 4 |
| FGF13 | C9orf24 | 0.6426 | 3 | 0 | 3 |
| FGF13 | TCEAL3 | 0.641496 | 4 | 0 | 4 |
| FGF13 | NR2F1-AS1 | 0.636333 | 5 | 0 | 4 |
| FGF13 | INMT | 0.635221 | 4 | 0 | 4 |
| FGF13 | TBX5 | 0.633963 | 3 | 0 | 3 |
| FGF13 | ARHGEF26 | 0.633512 | 7 | 0 | 6 |
| FGF13 | CAMK2G | 0.628206 | 3 | 0 | 3 |
| FGF13 | MRVI1 | 0.626486 | 4 | 0 | 3 |
| FGF13 | WIF1 | 0.624435 | 5 | 0 | 4 |
| FGF13 | CHRM3 | 0.622579 | 10 | 0 | 9 |
For details and further investigation, click here