| Full name: BCL2 like 11 | Alias Symbol: BOD|BimL|BimEL|BimS|BIM | ||
| Type: protein-coding gene | Cytoband: 2q13 | ||
| Entrez ID: 10018 | HGNC ID: HGNC:994 | Ensembl Gene: ENSG00000153094 | OMIM ID: 603827 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BCL2L11 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04210 | Apoptosis | |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) |
Expression of BCL2L11:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCL2L11 | 10018 | 225606_at | 0.4849 | 0.3667 | |
| GSE20347 | BCL2L11 | 10018 | 222343_at | 0.3478 | 0.0100 | |
| GSE23400 | BCL2L11 | 10018 | 222343_at | 0.0949 | 0.0117 | |
| GSE26886 | BCL2L11 | 10018 | 225606_at | 0.7118 | 0.0077 | |
| GSE29001 | BCL2L11 | 10018 | 222343_at | 0.6011 | 0.0369 | |
| GSE38129 | BCL2L11 | 10018 | 222343_at | 0.5187 | 0.0000 | |
| GSE45670 | BCL2L11 | 10018 | 225606_at | 0.3301 | 0.0353 | |
| GSE53622 | BCL2L11 | 10018 | 27651 | 0.6894 | 0.0000 | |
| GSE53624 | BCL2L11 | 10018 | 27651 | 0.6782 | 0.0000 | |
| GSE63941 | BCL2L11 | 10018 | 225606_at | 1.5514 | 0.0940 | |
| GSE77861 | BCL2L11 | 10018 | 1553096_s_at | -0.1401 | 0.3292 | |
| GSE97050 | BCL2L11 | 10018 | A_33_P3398526 | 0.9552 | 0.1496 | |
| SRP007169 | BCL2L11 | 10018 | RNAseq | 0.2005 | 0.6684 | |
| SRP008496 | BCL2L11 | 10018 | RNAseq | -1.2864 | 0.1254 | |
| SRP064894 | BCL2L11 | 10018 | RNAseq | 0.3308 | 0.1474 | |
| SRP133303 | BCL2L11 | 10018 | RNAseq | 0.6135 | 0.0032 | |
| SRP159526 | BCL2L11 | 10018 | RNAseq | 0.6389 | 0.0308 | |
| SRP193095 | BCL2L11 | 10018 | RNAseq | 0.2944 | 0.0382 | |
| SRP219564 | BCL2L11 | 10018 | RNAseq | -0.0289 | 0.9328 | |
| TCGA | BCL2L11 | 10018 | RNAseq | 0.2325 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
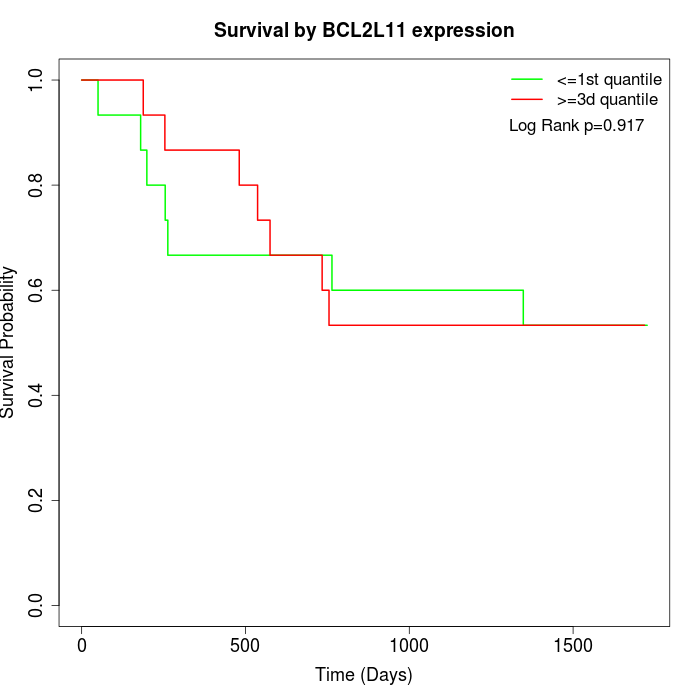
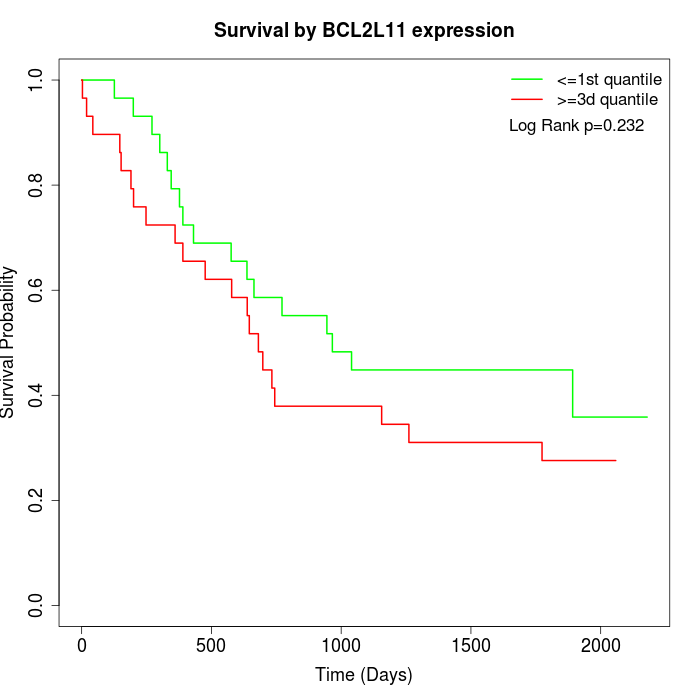
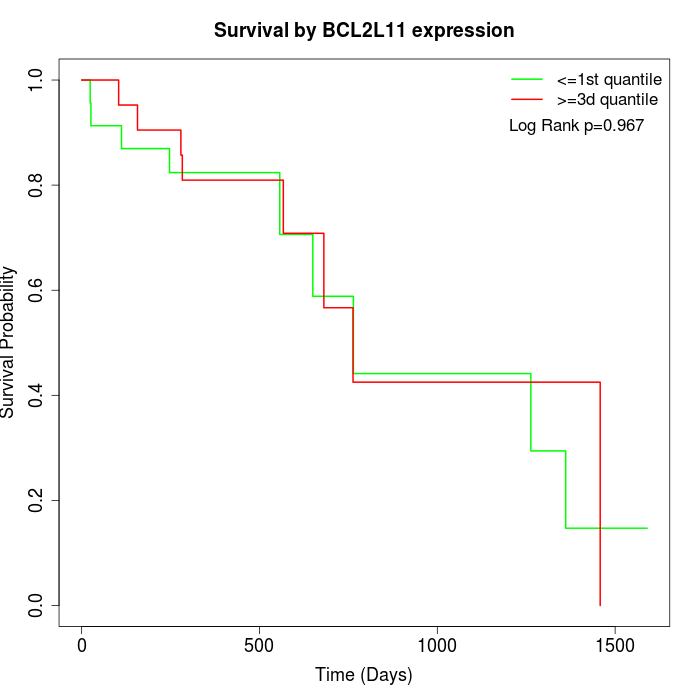
Survival by BCL2L11 expression:
Note: Click image to view full size file.
Copy number change of BCL2L11:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCL2L11 | 10018 | 4 | 2 | 24 | |
| GSE20123 | BCL2L11 | 10018 | 4 | 2 | 24 | |
| GSE43470 | BCL2L11 | 10018 | 3 | 1 | 39 | |
| GSE46452 | BCL2L11 | 10018 | 1 | 4 | 54 | |
| GSE47630 | BCL2L11 | 10018 | 7 | 0 | 33 | |
| GSE54993 | BCL2L11 | 10018 | 0 | 6 | 64 | |
| GSE54994 | BCL2L11 | 10018 | 10 | 0 | 43 | |
| GSE60625 | BCL2L11 | 10018 | 0 | 3 | 8 | |
| GSE74703 | BCL2L11 | 10018 | 3 | 1 | 32 | |
| GSE74704 | BCL2L11 | 10018 | 2 | 2 | 16 | |
| TCGA | BCL2L11 | 10018 | 31 | 5 | 60 |
Total number of gains: 65; Total number of losses: 26; Total Number of normals: 397.
Somatic mutations of BCL2L11:
Generating mutation plots.
Highly correlated genes for BCL2L11:
Showing top 20/374 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCL2L11 | GZMA | 0.854195 | 3 | 0 | 3 |
| BCL2L11 | SPOCK2 | 0.826525 | 3 | 0 | 3 |
| BCL2L11 | VASH1 | 0.80235 | 3 | 0 | 3 |
| BCL2L11 | SUSD3 | 0.776741 | 4 | 0 | 3 |
| BCL2L11 | KLRC3 | 0.745368 | 3 | 0 | 3 |
| BCL2L11 | ACAP1 | 0.736724 | 4 | 0 | 3 |
| BCL2L11 | ENPP5 | 0.728664 | 3 | 0 | 3 |
| BCL2L11 | SSBP4 | 0.719337 | 3 | 0 | 3 |
| BCL2L11 | XXYLT1 | 0.71782 | 3 | 0 | 3 |
| BCL2L11 | FAM78A | 0.715198 | 3 | 0 | 3 |
| BCL2L11 | DERL3 | 0.710592 | 3 | 0 | 3 |
| BCL2L11 | CDCA7 | 0.706039 | 4 | 0 | 4 |
| BCL2L11 | SGPL1 | 0.704932 | 3 | 0 | 3 |
| BCL2L11 | HAVCR2 | 0.697338 | 3 | 0 | 3 |
| BCL2L11 | LAPTM5 | 0.696507 | 4 | 0 | 3 |
| BCL2L11 | CCDC97 | 0.694023 | 4 | 0 | 4 |
| BCL2L11 | LPXN | 0.687762 | 3 | 0 | 3 |
| BCL2L11 | FUT8 | 0.684041 | 4 | 0 | 3 |
| BCL2L11 | RFXAP | 0.682878 | 4 | 0 | 4 |
| BCL2L11 | DHRSX | 0.682592 | 3 | 0 | 3 |
For details and further investigation, click here