| Full name: BCL9 transcription coactivator | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q21.2 | ||
| Entrez ID: 607 | HGNC ID: HGNC:1008 | Ensembl Gene: ENSG00000116128 | OMIM ID: 602597 |
| Drug and gene relationship at DGIdb | |||
Expression of BCL9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCL9 | 607 | 204129_at | 0.3423 | 0.1419 | |
| GSE20347 | BCL9 | 607 | 204129_at | 0.0960 | 0.1683 | |
| GSE23400 | BCL9 | 607 | 204129_at | -0.0059 | 0.8289 | |
| GSE26886 | BCL9 | 607 | 204129_at | 0.2108 | 0.0738 | |
| GSE29001 | BCL9 | 607 | 204129_at | 0.0079 | 0.9672 | |
| GSE38129 | BCL9 | 607 | 204129_at | 0.0129 | 0.8891 | |
| GSE45670 | BCL9 | 607 | 204129_at | -0.1382 | 0.2317 | |
| GSE53622 | BCL9 | 607 | 32528 | 0.1194 | 0.1836 | |
| GSE53624 | BCL9 | 607 | 32528 | 0.2250 | 0.0081 | |
| GSE63941 | BCL9 | 607 | 204129_at | 0.1457 | 0.5669 | |
| GSE77861 | BCL9 | 607 | 204129_at | 0.0316 | 0.8266 | |
| GSE97050 | BCL9 | 607 | A_23_P382602 | -0.3085 | 0.2874 | |
| SRP007169 | BCL9 | 607 | RNAseq | 0.6435 | 0.2046 | |
| SRP008496 | BCL9 | 607 | RNAseq | 0.9383 | 0.0288 | |
| SRP064894 | BCL9 | 607 | RNAseq | 0.0478 | 0.7738 | |
| SRP133303 | BCL9 | 607 | RNAseq | 0.0020 | 0.9888 | |
| SRP159526 | BCL9 | 607 | RNAseq | 0.4423 | 0.2178 | |
| SRP193095 | BCL9 | 607 | RNAseq | 0.1570 | 0.2334 | |
| SRP219564 | BCL9 | 607 | RNAseq | 0.0477 | 0.9018 | |
| TCGA | BCL9 | 607 | RNAseq | -0.0644 | 0.2409 |
Upregulated datasets: 0; Downregulated datasets: 0.
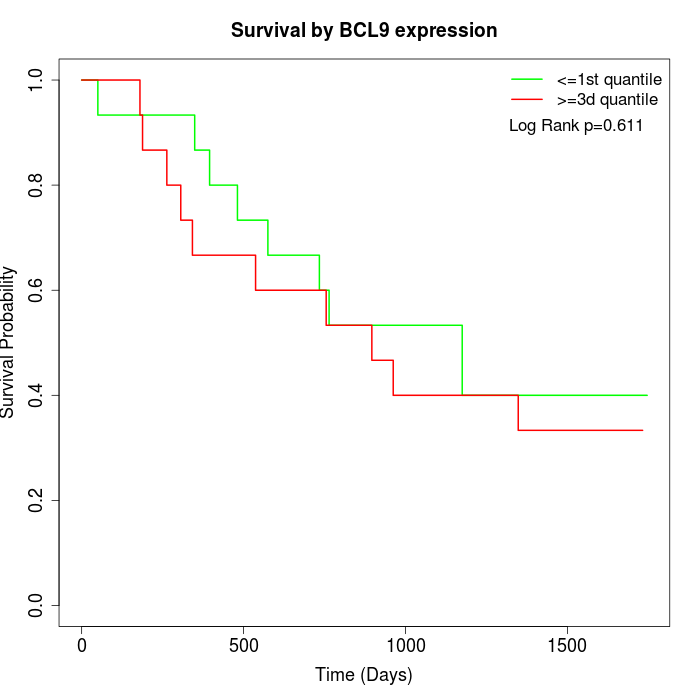
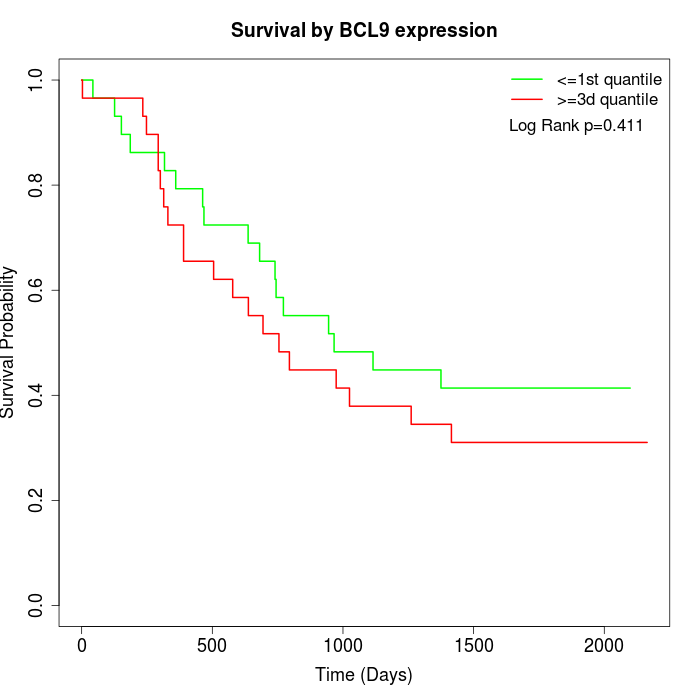
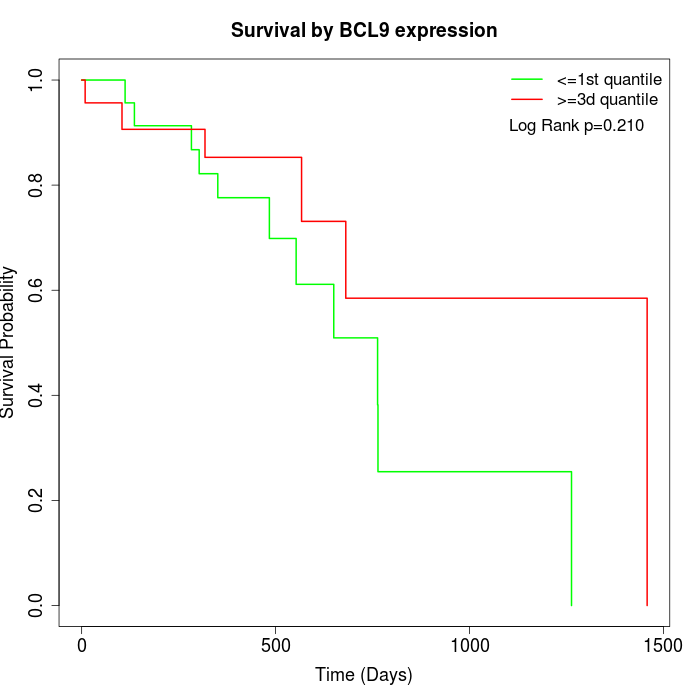
Survival by BCL9 expression:
Note: Click image to view full size file.
Copy number change of BCL9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCL9 | 607 | 12 | 0 | 18 | |
| GSE20123 | BCL9 | 607 | 12 | 0 | 18 | |
| GSE43470 | BCL9 | 607 | 5 | 1 | 37 | |
| GSE46452 | BCL9 | 607 | 2 | 1 | 56 | |
| GSE47630 | BCL9 | 607 | 12 | 2 | 26 | |
| GSE54993 | BCL9 | 607 | 0 | 2 | 68 | |
| GSE54994 | BCL9 | 607 | 15 | 0 | 38 | |
| GSE60625 | BCL9 | 607 | 0 | 0 | 11 | |
| GSE74703 | BCL9 | 607 | 5 | 1 | 30 | |
| GSE74704 | BCL9 | 607 | 6 | 0 | 14 | |
| TCGA | BCL9 | 607 | 33 | 15 | 48 |
Total number of gains: 102; Total number of losses: 22; Total Number of normals: 364.
Somatic mutations of BCL9:
Generating mutation plots.
Highly correlated genes for BCL9:
Showing top 20/412 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCL9 | ERF | 0.82013 | 3 | 0 | 3 |
| BCL9 | ZNF787 | 0.780626 | 3 | 0 | 3 |
| BCL9 | RB1CC1 | 0.77073 | 3 | 0 | 3 |
| BCL9 | PRR12 | 0.761556 | 3 | 0 | 3 |
| BCL9 | INPP5E | 0.754663 | 3 | 0 | 3 |
| BCL9 | PKNOX1 | 0.747969 | 3 | 0 | 3 |
| BCL9 | PET117 | 0.743949 | 3 | 0 | 3 |
| BCL9 | GNA11 | 0.743886 | 4 | 0 | 4 |
| BCL9 | CLCN7 | 0.737484 | 3 | 0 | 3 |
| BCL9 | TRIM47 | 0.73431 | 3 | 0 | 3 |
| BCL9 | SKI | 0.732926 | 3 | 0 | 3 |
| BCL9 | ZMIZ2 | 0.731811 | 3 | 0 | 3 |
| BCL9 | MED1 | 0.727834 | 3 | 0 | 3 |
| BCL9 | EVI5L | 0.726393 | 3 | 0 | 3 |
| BCL9 | ZBTB37 | 0.721206 | 4 | 0 | 3 |
| BCL9 | TET1 | 0.718001 | 4 | 0 | 4 |
| BCL9 | STAT5B | 0.716374 | 3 | 0 | 3 |
| BCL9 | ZNF391 | 0.716259 | 4 | 0 | 3 |
| BCL9 | DEXI | 0.716247 | 3 | 0 | 3 |
| BCL9 | LRRC58 | 0.707876 | 3 | 0 | 3 |
For details and further investigation, click here