| Full name: BCR activator of RhoGEF and GTPase | Alias Symbol: D22S662|CML|PHL|ALL | ||
| Type: protein-coding gene | Cytoband: 22q11.23 | ||
| Entrez ID: 613 | HGNC ID: HGNC:1014 | Ensembl Gene: ENSG00000186716 | OMIM ID: 151410 |
| Drug and gene relationship at DGIdb | |||
BCR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05200 | Pathways in cancer | |
| hsa05220 | Chronic myeloid leukemia |
Expression of BCR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BCR | 613 | 202315_s_at | 0.5099 | 0.4141 | |
| GSE20347 | BCR | 613 | 202315_s_at | -0.3646 | 0.0696 | |
| GSE23400 | BCR | 613 | 217223_s_at | -0.2345 | 0.0009 | |
| GSE26886 | BCR | 613 | 202315_s_at | -0.7939 | 0.0281 | |
| GSE29001 | BCR | 613 | 202315_s_at | -0.6078 | 0.0956 | |
| GSE38129 | BCR | 613 | 202315_s_at | -0.0207 | 0.9236 | |
| GSE45670 | BCR | 613 | 202315_s_at | 0.3230 | 0.1115 | |
| GSE53622 | BCR | 613 | 65051 | -2.3136 | 0.0000 | |
| GSE53624 | BCR | 613 | 50389 | -0.3557 | 0.0000 | |
| GSE63941 | BCR | 613 | 202315_s_at | 0.2496 | 0.7059 | |
| GSE77861 | BCR | 613 | 226602_s_at | 0.3610 | 0.0353 | |
| GSE97050 | BCR | 613 | A_24_P127235 | 0.2220 | 0.4974 | |
| SRP007169 | BCR | 613 | RNAseq | -0.6888 | 0.1140 | |
| SRP008496 | BCR | 613 | RNAseq | -0.8071 | 0.0012 | |
| SRP064894 | BCR | 613 | RNAseq | -0.4281 | 0.0271 | |
| SRP133303 | BCR | 613 | RNAseq | -0.3013 | 0.1194 | |
| SRP159526 | BCR | 613 | RNAseq | -0.2176 | 0.3219 | |
| SRP193095 | BCR | 613 | RNAseq | 0.0247 | 0.8661 | |
| SRP219564 | BCR | 613 | RNAseq | -0.1810 | 0.6110 | |
| TCGA | BCR | 613 | RNAseq | -0.0182 | 0.7235 |
Upregulated datasets: 0; Downregulated datasets: 1.
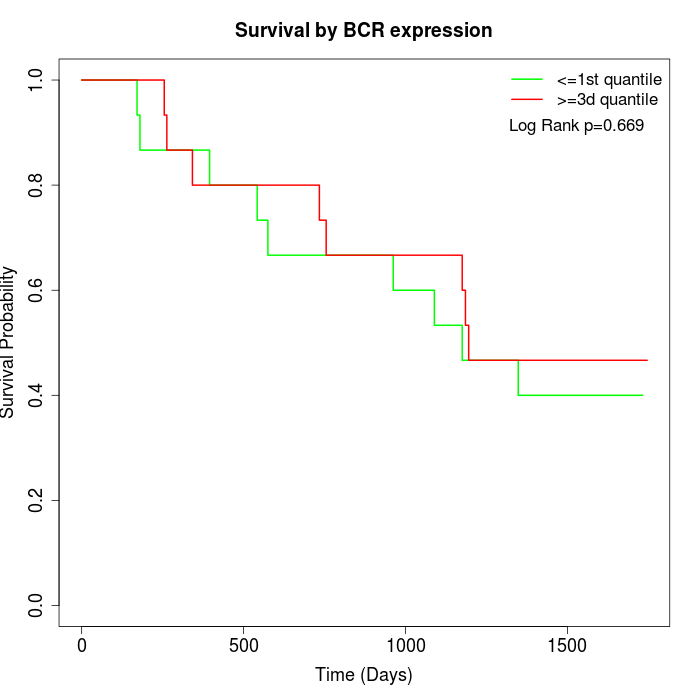
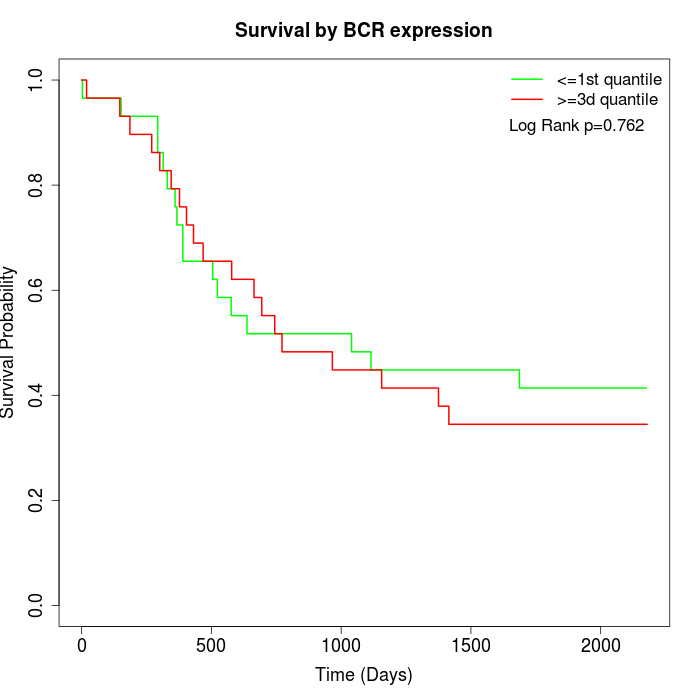
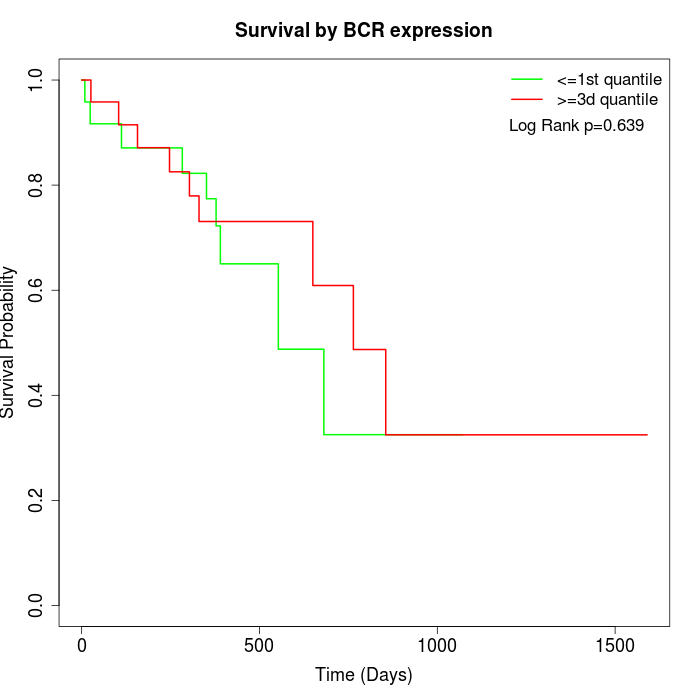
Survival by BCR expression:
Note: Click image to view full size file.
Copy number change of BCR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BCR | 613 | 5 | 7 | 18 | |
| GSE20123 | BCR | 613 | 5 | 6 | 19 | |
| GSE43470 | BCR | 613 | 3 | 6 | 34 | |
| GSE46452 | BCR | 613 | 32 | 1 | 26 | |
| GSE47630 | BCR | 613 | 8 | 5 | 27 | |
| GSE54993 | BCR | 613 | 3 | 6 | 61 | |
| GSE54994 | BCR | 613 | 11 | 7 | 35 | |
| GSE60625 | BCR | 613 | 5 | 0 | 6 | |
| GSE74703 | BCR | 613 | 3 | 5 | 28 | |
| GSE74704 | BCR | 613 | 2 | 3 | 15 | |
| TCGA | BCR | 613 | 27 | 16 | 53 |
Total number of gains: 104; Total number of losses: 62; Total Number of normals: 322.
Somatic mutations of BCR:
Generating mutation plots.
Highly correlated genes for BCR:
Showing top 20/83 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BCR | CHEK2 | 0.706298 | 3 | 0 | 3 |
| BCR | KRTCAP3 | 0.672928 | 3 | 0 | 3 |
| BCR | BIN3 | 0.660197 | 4 | 0 | 3 |
| BCR | EIF1AD | 0.651319 | 3 | 0 | 3 |
| BCR | RANGAP1 | 0.633484 | 4 | 0 | 3 |
| BCR | F11R | 0.631052 | 6 | 0 | 4 |
| BCR | FOXRED1 | 0.631016 | 4 | 0 | 3 |
| BCR | SERINC2 | 0.614551 | 4 | 0 | 3 |
| BCR | RAD9A | 0.61422 | 4 | 0 | 3 |
| BCR | FKBP1B | 0.610936 | 3 | 0 | 3 |
| BCR | WDR34 | 0.591726 | 4 | 0 | 3 |
| BCR | RNPEP | 0.591335 | 7 | 0 | 5 |
| BCR | IRF6 | 0.590249 | 5 | 0 | 3 |
| BCR | HM13 | 0.59019 | 6 | 0 | 3 |
| BCR | CHRNA5 | 0.584784 | 3 | 0 | 3 |
| BCR | SLC37A2 | 0.582393 | 5 | 0 | 3 |
| BCR | EHD4 | 0.581136 | 7 | 0 | 5 |
| BCR | TMPRSS4 | 0.576671 | 6 | 0 | 4 |
| BCR | TOMM34 | 0.575498 | 5 | 0 | 3 |
| BCR | DDX52 | 0.575001 | 4 | 0 | 3 |
For details and further investigation, click here