| Full name: BRCA1 associated protein | Alias Symbol: BRAP2|RNF52|IMP | ||
| Type: protein-coding gene | Cytoband: 12q24.12 | ||
| Entrez ID: 8315 | HGNC ID: HGNC:1099 | Ensembl Gene: ENSG00000089234 | OMIM ID: 604986 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BRAP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway |
Expression of BRAP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BRAP | 8315 | 213473_at | -0.1280 | 0.8016 | |
| GSE20347 | BRAP | 8315 | 213473_at | 0.0086 | 0.9572 | |
| GSE23400 | BRAP | 8315 | 209923_s_at | 0.1371 | 0.0023 | |
| GSE26886 | BRAP | 8315 | 209923_s_at | 0.4789 | 0.0001 | |
| GSE29001 | BRAP | 8315 | 213473_at | 0.1680 | 0.4749 | |
| GSE38129 | BRAP | 8315 | 213473_at | 0.0422 | 0.7667 | |
| GSE45670 | BRAP | 8315 | 213473_at | -0.0481 | 0.7691 | |
| GSE53622 | BRAP | 8315 | 60493 | 0.2219 | 0.0002 | |
| GSE53624 | BRAP | 8315 | 60493 | 0.1949 | 0.0112 | |
| GSE63941 | BRAP | 8315 | 213473_at | -0.9552 | 0.0067 | |
| GSE77861 | BRAP | 8315 | 209923_s_at | 0.1349 | 0.1552 | |
| GSE97050 | BRAP | 8315 | A_23_P53614 | -0.1297 | 0.5830 | |
| SRP007169 | BRAP | 8315 | RNAseq | 0.3439 | 0.3545 | |
| SRP008496 | BRAP | 8315 | RNAseq | 0.4544 | 0.0601 | |
| SRP064894 | BRAP | 8315 | RNAseq | -0.0436 | 0.7034 | |
| SRP133303 | BRAP | 8315 | RNAseq | 0.2182 | 0.0424 | |
| SRP159526 | BRAP | 8315 | RNAseq | 0.1434 | 0.4536 | |
| SRP193095 | BRAP | 8315 | RNAseq | -0.0718 | 0.2641 | |
| SRP219564 | BRAP | 8315 | RNAseq | -0.2235 | 0.3123 | |
| TCGA | BRAP | 8315 | RNAseq | 0.0021 | 0.9693 |
Upregulated datasets: 0; Downregulated datasets: 0.
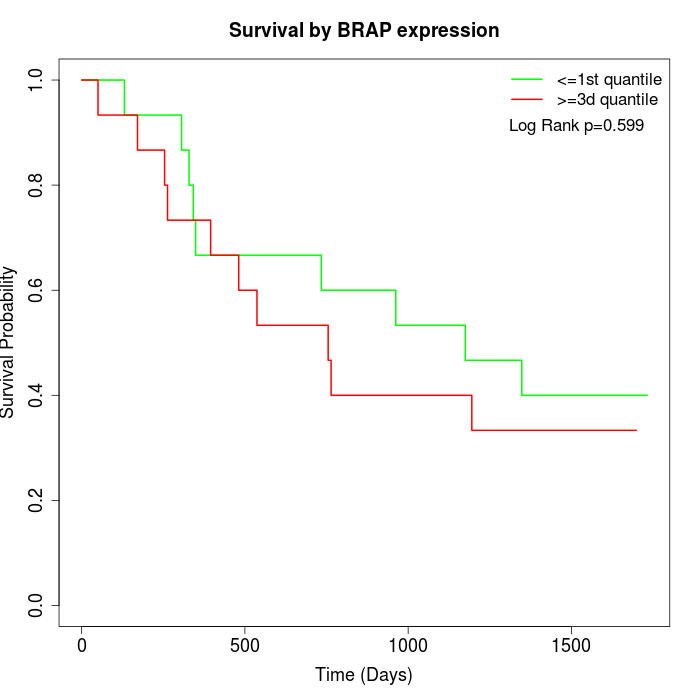
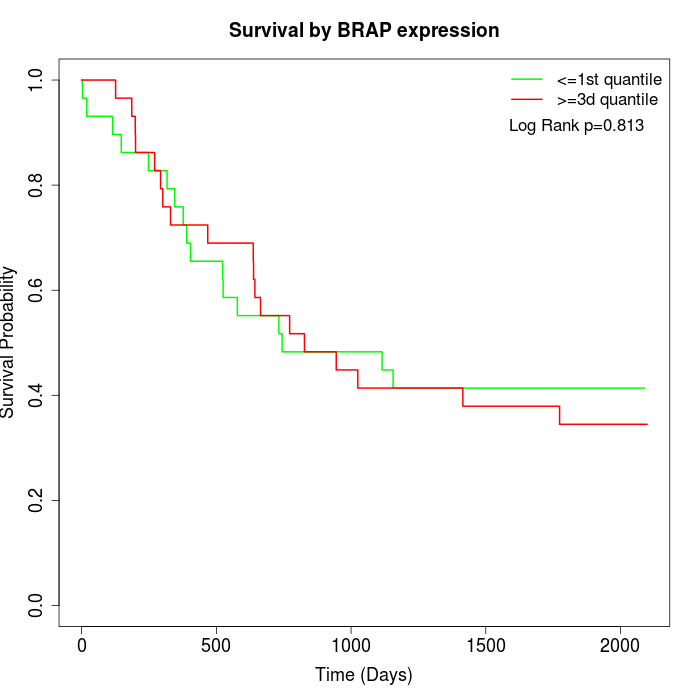
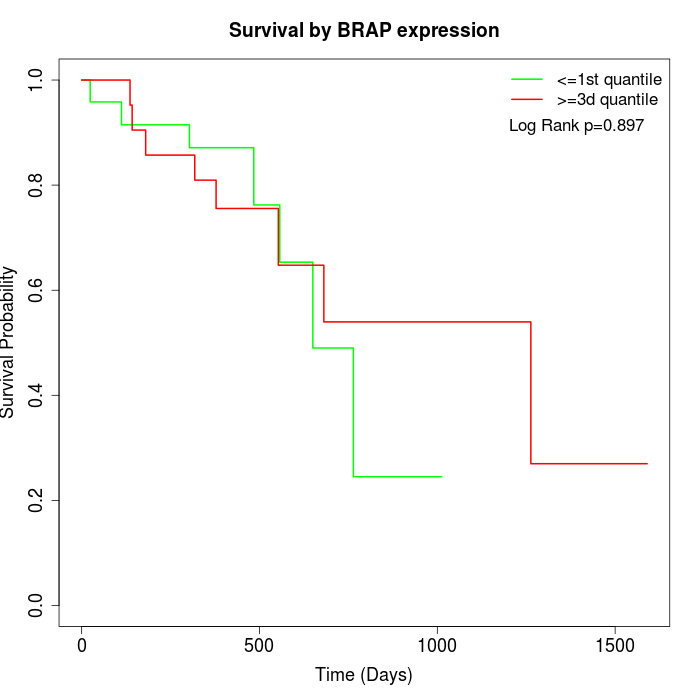
Survival by BRAP expression:
Note: Click image to view full size file.
Copy number change of BRAP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BRAP | 8315 | 5 | 3 | 22 | |
| GSE20123 | BRAP | 8315 | 5 | 3 | 22 | |
| GSE43470 | BRAP | 8315 | 2 | 1 | 40 | |
| GSE46452 | BRAP | 8315 | 9 | 1 | 49 | |
| GSE47630 | BRAP | 8315 | 9 | 3 | 28 | |
| GSE54993 | BRAP | 8315 | 0 | 5 | 65 | |
| GSE54994 | BRAP | 8315 | 4 | 2 | 47 | |
| GSE60625 | BRAP | 8315 | 0 | 0 | 11 | |
| GSE74703 | BRAP | 8315 | 2 | 0 | 34 | |
| GSE74704 | BRAP | 8315 | 2 | 2 | 16 | |
| TCGA | BRAP | 8315 | 22 | 11 | 63 |
Total number of gains: 60; Total number of losses: 31; Total Number of normals: 397.
Somatic mutations of BRAP:
Generating mutation plots.
Highly correlated genes for BRAP:
Showing top 20/483 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BRAP | SCAF8 | 0.8217 | 3 | 0 | 3 |
| BRAP | SLC35A5 | 0.799447 | 3 | 0 | 3 |
| BRAP | ZCCHC8 | 0.792413 | 3 | 0 | 3 |
| BRAP | LZIC | 0.787489 | 3 | 0 | 3 |
| BRAP | PNKD | 0.780792 | 3 | 0 | 3 |
| BRAP | RIOK1 | 0.772441 | 3 | 0 | 3 |
| BRAP | NPHP3 | 0.770055 | 3 | 0 | 3 |
| BRAP | CHIC1 | 0.761897 | 3 | 0 | 3 |
| BRAP | ALKBH6 | 0.760132 | 3 | 0 | 3 |
| BRAP | ZNF317 | 0.745829 | 3 | 0 | 3 |
| BRAP | DMAP1 | 0.740236 | 3 | 0 | 3 |
| BRAP | CTPS2 | 0.737091 | 3 | 0 | 3 |
| BRAP | CAMTA1 | 0.735435 | 3 | 0 | 3 |
| BRAP | PLEKHB2 | 0.734033 | 3 | 0 | 3 |
| BRAP | KLHL15 | 0.73364 | 3 | 0 | 3 |
| BRAP | DEDD2 | 0.73219 | 3 | 0 | 3 |
| BRAP | MOB3C | 0.730373 | 3 | 0 | 3 |
| BRAP | PAK1 | 0.729728 | 3 | 0 | 3 |
| BRAP | HNRNPUL2 | 0.727327 | 4 | 0 | 3 |
| BRAP | RSRC2 | 0.72551 | 5 | 0 | 3 |
For details and further investigation, click here