| Full name: cyclin L2 | Alias Symbol: ania-6b|PCEE|SB138|HLA-ISO|CCNS | ||
| Type: protein-coding gene | Cytoband: 1p36.33 | ||
| Entrez ID: 81669 | HGNC ID: HGNC:20570 | Ensembl Gene: ENSG00000221978 | OMIM ID: 613482 |
| Drug and gene relationship at DGIdb | |||
Expression of CCNL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNL2 | 81669 | 222999_s_at | 0.2369 | 0.6178 | |
| GSE20347 | CCNL2 | 81669 | 221427_s_at | 0.2275 | 0.3162 | |
| GSE23400 | CCNL2 | 81669 | 221427_s_at | 0.3229 | 0.0001 | |
| GSE26886 | CCNL2 | 81669 | 222999_s_at | 0.4115 | 0.1631 | |
| GSE29001 | CCNL2 | 81669 | 221427_s_at | 0.0497 | 0.9122 | |
| GSE38129 | CCNL2 | 81669 | 221427_s_at | 0.2811 | 0.1903 | |
| GSE45670 | CCNL2 | 81669 | 222999_s_at | 0.1233 | 0.5513 | |
| GSE53622 | CCNL2 | 81669 | 23047 | 0.1757 | 0.0165 | |
| GSE53624 | CCNL2 | 81669 | 23047 | 0.2341 | 0.0152 | |
| GSE63941 | CCNL2 | 81669 | 222999_s_at | -0.3761 | 0.5057 | |
| GSE77861 | CCNL2 | 81669 | 222999_s_at | 0.4103 | 0.3362 | |
| GSE97050 | CCNL2 | 81669 | A_23_P217899 | 0.0535 | 0.7935 | |
| SRP007169 | CCNL2 | 81669 | RNAseq | -0.9433 | 0.0102 | |
| SRP008496 | CCNL2 | 81669 | RNAseq | -0.5799 | 0.0381 | |
| SRP064894 | CCNL2 | 81669 | RNAseq | 0.6156 | 0.0007 | |
| SRP133303 | CCNL2 | 81669 | RNAseq | 0.0140 | 0.9388 | |
| SRP159526 | CCNL2 | 81669 | RNAseq | 0.1346 | 0.7962 | |
| SRP193095 | CCNL2 | 81669 | RNAseq | 0.0797 | 0.5012 | |
| SRP219564 | CCNL2 | 81669 | RNAseq | -0.0224 | 0.9350 | |
| TCGA | CCNL2 | 81669 | RNAseq | -0.0018 | 0.9726 |
Upregulated datasets: 0; Downregulated datasets: 0.
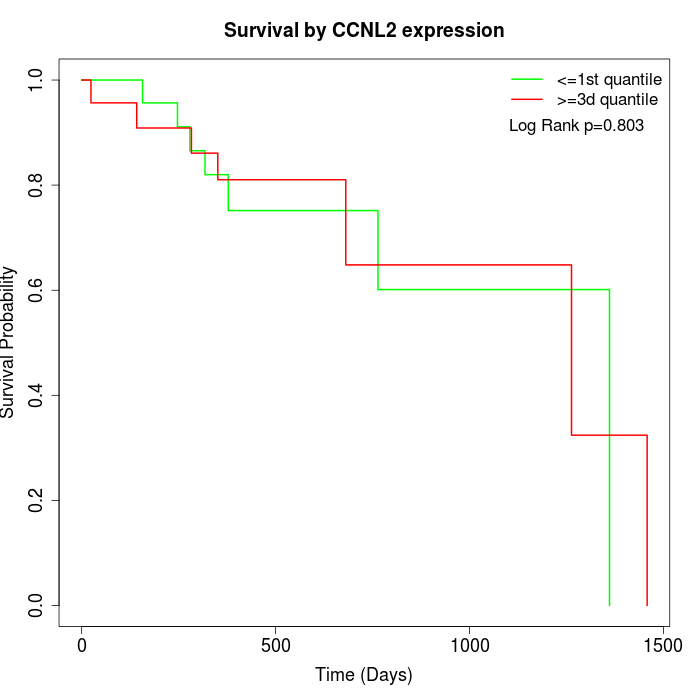
Survival by CCNL2 expression:
Note: Click image to view full size file.
Copy number change of CCNL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNL2 | 81669 | 3 | 7 | 20 | |
| GSE20123 | CCNL2 | 81669 | 3 | 6 | 21 | |
| GSE43470 | CCNL2 | 81669 | 3 | 5 | 35 | |
| GSE46452 | CCNL2 | 81669 | 7 | 1 | 51 | |
| GSE47630 | CCNL2 | 81669 | 8 | 4 | 28 | |
| GSE54993 | CCNL2 | 81669 | 2 | 2 | 66 | |
| GSE54994 | CCNL2 | 81669 | 15 | 2 | 36 | |
| GSE60625 | CCNL2 | 81669 | 0 | 0 | 11 | |
| GSE74703 | CCNL2 | 81669 | 3 | 3 | 30 | |
| GSE74704 | CCNL2 | 81669 | 3 | 0 | 17 | |
| TCGA | CCNL2 | 81669 | 14 | 20 | 62 |
Total number of gains: 61; Total number of losses: 50; Total Number of normals: 377.
Somatic mutations of CCNL2:
Generating mutation plots.
Highly correlated genes for CCNL2:
Showing top 20/114 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNL2 | RPA4 | 0.731804 | 3 | 0 | 3 |
| CCNL2 | ZNF511 | 0.72115 | 3 | 0 | 3 |
| CCNL2 | TIGD7 | 0.714028 | 3 | 0 | 3 |
| CCNL2 | CDK5R2 | 0.713735 | 3 | 0 | 3 |
| CCNL2 | ZBTB48 | 0.686819 | 9 | 0 | 8 |
| CCNL2 | CABP4 | 0.681608 | 3 | 0 | 3 |
| CCNL2 | MBTD1 | 0.679169 | 4 | 0 | 3 |
| CCNL2 | POLL | 0.666175 | 4 | 0 | 3 |
| CCNL2 | ADD2 | 0.664755 | 4 | 0 | 3 |
| CCNL2 | CDK12 | 0.662825 | 4 | 0 | 3 |
| CCNL2 | CES3 | 0.661646 | 5 | 0 | 5 |
| CCNL2 | ATG16L1 | 0.659268 | 4 | 0 | 3 |
| CCNL2 | TRIM41 | 0.657593 | 4 | 0 | 4 |
| CCNL2 | PCYOX1L | 0.655922 | 3 | 0 | 3 |
| CCNL2 | PYGO2 | 0.65575 | 3 | 0 | 3 |
| CCNL2 | CCDC130 | 0.650906 | 9 | 0 | 7 |
| CCNL2 | TNFAIP8L1 | 0.649184 | 4 | 0 | 4 |
| CCNL2 | GTF3C5 | 0.638859 | 5 | 0 | 4 |
| CCNL2 | RXRB | 0.638728 | 4 | 0 | 3 |
| CCNL2 | CNOT3 | 0.635278 | 5 | 0 | 3 |
For details and further investigation, click here