| Full name: autophagy related 16 like 1 | Alias Symbol: WDR30|FLJ10035|ATG16A | ||
| Type: protein-coding gene | Cytoband: 2q37.1 | ||
| Entrez ID: 55054 | HGNC ID: HGNC:21498 | Ensembl Gene: ENSG00000085978 | OMIM ID: 610767 |
| Drug and gene relationship at DGIdb | |||
Expression of ATG16L1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATG16L1 | 55054 | 232612_s_at | 0.3527 | 0.2110 | |
| GSE20347 | ATG16L1 | 55054 | 220521_s_at | -0.0819 | 0.4286 | |
| GSE23400 | ATG16L1 | 55054 | 220521_s_at | -0.0328 | 0.3559 | |
| GSE26886 | ATG16L1 | 55054 | 232612_s_at | -0.0749 | 0.7179 | |
| GSE29001 | ATG16L1 | 55054 | 220521_s_at | -0.1368 | 0.3338 | |
| GSE38129 | ATG16L1 | 55054 | 220521_s_at | -0.1139 | 0.4568 | |
| GSE45670 | ATG16L1 | 55054 | 232612_s_at | 0.1588 | 0.2897 | |
| GSE53622 | ATG16L1 | 55054 | 38791 | 0.0212 | 0.7871 | |
| GSE53624 | ATG16L1 | 55054 | 38791 | 0.0935 | 0.1006 | |
| GSE63941 | ATG16L1 | 55054 | 232612_s_at | -0.1151 | 0.7881 | |
| GSE77861 | ATG16L1 | 55054 | 232612_s_at | 0.2599 | 0.0868 | |
| GSE97050 | ATG16L1 | 55054 | A_33_P3352134 | 0.0913 | 0.7206 | |
| SRP007169 | ATG16L1 | 55054 | RNAseq | 0.0803 | 0.8419 | |
| SRP008496 | ATG16L1 | 55054 | RNAseq | 0.1669 | 0.4456 | |
| SRP064894 | ATG16L1 | 55054 | RNAseq | -0.0556 | 0.6429 | |
| SRP133303 | ATG16L1 | 55054 | RNAseq | 0.2078 | 0.1587 | |
| SRP159526 | ATG16L1 | 55054 | RNAseq | 0.0309 | 0.8849 | |
| SRP193095 | ATG16L1 | 55054 | RNAseq | 0.0498 | 0.6797 | |
| SRP219564 | ATG16L1 | 55054 | RNAseq | 0.0052 | 0.9829 | |
| TCGA | ATG16L1 | 55054 | RNAseq | -0.1263 | 0.0249 |
Upregulated datasets: 0; Downregulated datasets: 0.
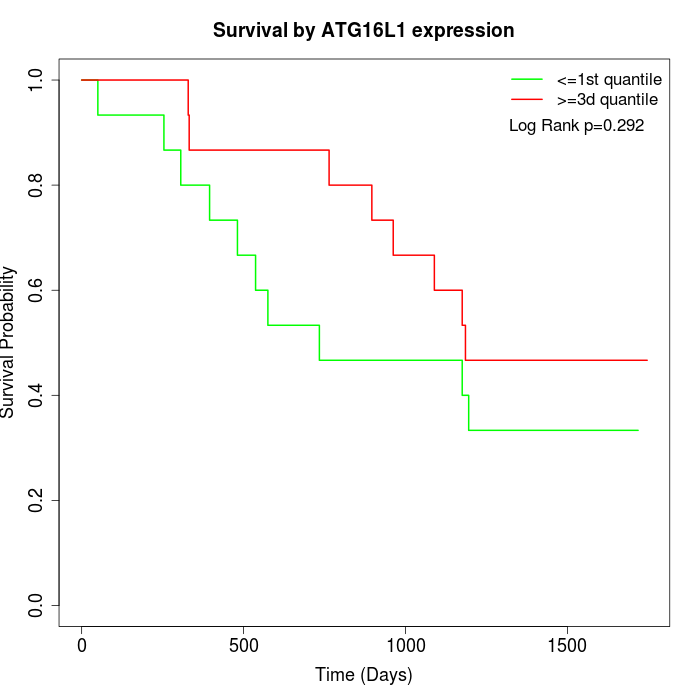
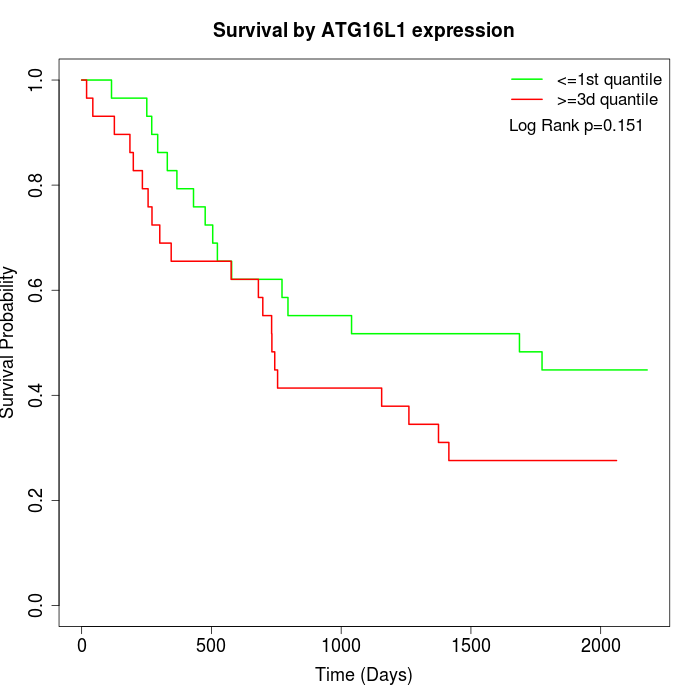
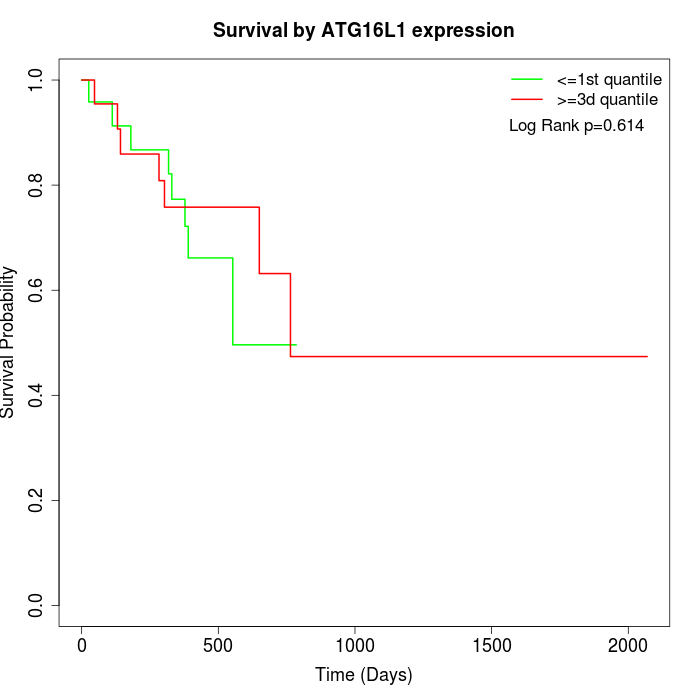
Survival by ATG16L1 expression:
Note: Click image to view full size file.
Copy number change of ATG16L1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATG16L1 | 55054 | 1 | 12 | 17 | |
| GSE20123 | ATG16L1 | 55054 | 1 | 12 | 17 | |
| GSE43470 | ATG16L1 | 55054 | 1 | 9 | 33 | |
| GSE46452 | ATG16L1 | 55054 | 0 | 5 | 54 | |
| GSE47630 | ATG16L1 | 55054 | 4 | 5 | 31 | |
| GSE54993 | ATG16L1 | 55054 | 3 | 2 | 65 | |
| GSE54994 | ATG16L1 | 55054 | 6 | 10 | 37 | |
| GSE60625 | ATG16L1 | 55054 | 0 | 3 | 8 | |
| GSE74703 | ATG16L1 | 55054 | 1 | 7 | 28 | |
| GSE74704 | ATG16L1 | 55054 | 1 | 5 | 14 | |
| TCGA | ATG16L1 | 55054 | 12 | 28 | 56 |
Total number of gains: 30; Total number of losses: 98; Total Number of normals: 360.
Somatic mutations of ATG16L1:
Generating mutation plots.
Highly correlated genes for ATG16L1:
Showing top 20/112 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATG16L1 | KCNAB3 | 0.734069 | 3 | 0 | 3 |
| ATG16L1 | CDX2 | 0.724486 | 3 | 0 | 3 |
| ATG16L1 | MAT1A | 0.72114 | 3 | 0 | 3 |
| ATG16L1 | KIF4A | 0.721105 | 3 | 0 | 3 |
| ATG16L1 | CCL26 | 0.720793 | 3 | 0 | 3 |
| ATG16L1 | PLEC | 0.713895 | 4 | 0 | 3 |
| ATG16L1 | RASL11B | 0.71277 | 3 | 0 | 3 |
| ATG16L1 | GPRIN1 | 0.710377 | 3 | 0 | 3 |
| ATG16L1 | FOLR3 | 0.690682 | 3 | 0 | 3 |
| ATG16L1 | ENTPD5 | 0.685029 | 3 | 0 | 3 |
| ATG16L1 | NPPA | 0.682113 | 3 | 0 | 3 |
| ATG16L1 | AQP6 | 0.681704 | 3 | 0 | 3 |
| ATG16L1 | CLPB | 0.671511 | 4 | 0 | 3 |
| ATG16L1 | NPEPL1 | 0.669792 | 4 | 0 | 3 |
| ATG16L1 | ZNF283 | 0.667535 | 3 | 0 | 3 |
| ATG16L1 | HMBS | 0.663321 | 3 | 0 | 3 |
| ATG16L1 | RECQL4 | 0.662344 | 3 | 0 | 3 |
| ATG16L1 | CCNL2 | 0.659268 | 4 | 0 | 3 |
| ATG16L1 | VAV2 | 0.658137 | 4 | 0 | 3 |
| ATG16L1 | FAM189B | 0.656343 | 3 | 0 | 3 |
For details and further investigation, click here