| Full name: cyclin T2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q21.3 | ||
| Entrez ID: 905 | HGNC ID: HGNC:1600 | Ensembl Gene: ENSG00000082258 | OMIM ID: 603862 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CCNT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNT2 | 905 | 213743_at | 0.3078 | 0.3870 | |
| GSE20347 | CCNT2 | 905 | 213743_at | -0.1906 | 0.2002 | |
| GSE23400 | CCNT2 | 905 | 213743_at | 0.1695 | 0.0319 | |
| GSE26886 | CCNT2 | 905 | 213743_at | 0.3058 | 0.3890 | |
| GSE29001 | CCNT2 | 905 | 213743_at | 0.0397 | 0.8882 | |
| GSE38129 | CCNT2 | 905 | 213743_at | 0.0169 | 0.9228 | |
| GSE45670 | CCNT2 | 905 | 213743_at | 0.0819 | 0.6588 | |
| GSE53622 | CCNT2 | 905 | 44447 | 0.4758 | 0.0000 | |
| GSE53624 | CCNT2 | 905 | 44447 | 0.3313 | 0.0001 | |
| GSE63941 | CCNT2 | 905 | 213743_at | -0.1889 | 0.6944 | |
| GSE77861 | CCNT2 | 905 | 213743_at | 0.3198 | 0.1227 | |
| GSE97050 | CCNT2 | 905 | A_24_P174341 | 0.0157 | 0.9519 | |
| SRP007169 | CCNT2 | 905 | RNAseq | -0.3412 | 0.3892 | |
| SRP008496 | CCNT2 | 905 | RNAseq | -0.1206 | 0.5753 | |
| SRP064894 | CCNT2 | 905 | RNAseq | -0.0304 | 0.9086 | |
| SRP133303 | CCNT2 | 905 | RNAseq | 0.3132 | 0.0516 | |
| SRP159526 | CCNT2 | 905 | RNAseq | -0.0163 | 0.9326 | |
| SRP193095 | CCNT2 | 905 | RNAseq | -0.1866 | 0.0198 | |
| SRP219564 | CCNT2 | 905 | RNAseq | -0.2901 | 0.3638 | |
| TCGA | CCNT2 | 905 | RNAseq | -0.0752 | 0.1394 |
Upregulated datasets: 0; Downregulated datasets: 0.
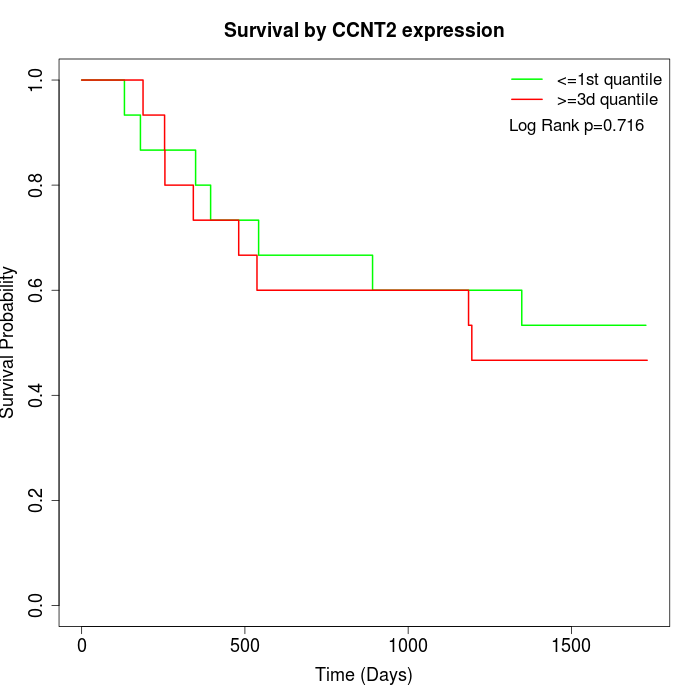
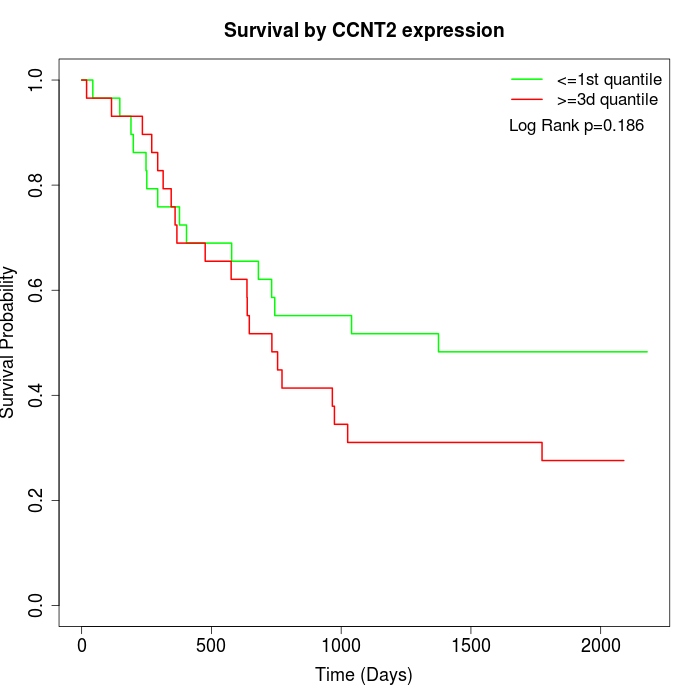
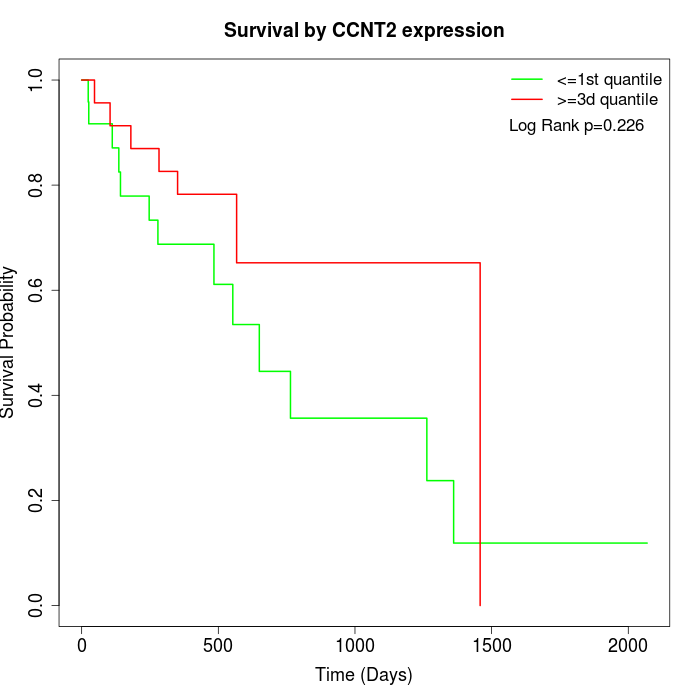
Survival by CCNT2 expression:
Note: Click image to view full size file.
Copy number change of CCNT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNT2 | 905 | 2 | 4 | 24 | |
| GSE20123 | CCNT2 | 905 | 2 | 4 | 24 | |
| GSE43470 | CCNT2 | 905 | 1 | 1 | 41 | |
| GSE46452 | CCNT2 | 905 | 1 | 4 | 54 | |
| GSE47630 | CCNT2 | 905 | 5 | 2 | 33 | |
| GSE54993 | CCNT2 | 905 | 0 | 6 | 64 | |
| GSE54994 | CCNT2 | 905 | 9 | 0 | 44 | |
| GSE60625 | CCNT2 | 905 | 0 | 3 | 8 | |
| GSE74703 | CCNT2 | 905 | 1 | 1 | 34 | |
| GSE74704 | CCNT2 | 905 | 1 | 1 | 18 | |
| TCGA | CCNT2 | 905 | 22 | 10 | 64 |
Total number of gains: 44; Total number of losses: 36; Total Number of normals: 408.
Somatic mutations of CCNT2:
Generating mutation plots.
Highly correlated genes for CCNT2:
Showing top 20/50 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNT2 | CCDC142 | 0.677092 | 3 | 0 | 3 |
| CCNT2 | TTPAL | 0.670799 | 3 | 0 | 3 |
| CCNT2 | ZNF302 | 0.659009 | 5 | 0 | 4 |
| CCNT2 | RWDD3 | 0.619024 | 4 | 0 | 4 |
| CCNT2 | WDPCP | 0.61494 | 4 | 0 | 3 |
| CCNT2 | DYNC2LI1 | 0.570619 | 4 | 0 | 3 |
| CCNT2 | ETAA1 | 0.564753 | 8 | 0 | 5 |
| CCNT2 | ZC3H8 | 0.563772 | 6 | 0 | 4 |
| CCNT2 | C5orf30 | 0.558043 | 4 | 0 | 3 |
| CCNT2 | FUT1 | 0.557362 | 4 | 0 | 3 |
| CCNT2 | TRMT13 | 0.554276 | 4 | 0 | 3 |
| CCNT2 | ITGA6 | 0.553617 | 4 | 0 | 3 |
| CCNT2 | PIGG | 0.553574 | 6 | 0 | 4 |
| CCNT2 | PM20D2 | 0.552976 | 3 | 0 | 3 |
| CCNT2 | CUL4A | 0.549441 | 8 | 0 | 5 |
| CCNT2 | TTC30A | 0.547355 | 6 | 0 | 4 |
| CCNT2 | THAP9-AS1 | 0.54329 | 4 | 0 | 3 |
| CCNT2 | KIF14 | 0.540126 | 4 | 0 | 3 |
| CCNT2 | MAP2 | 0.539458 | 4 | 0 | 3 |
| CCNT2 | ZBTB41 | 0.537966 | 6 | 0 | 5 |
For details and further investigation, click here