| Full name: GRB2 binding adaptor protein, transmembrane | Alias Symbol: FLJ33641 | ||
| Type: protein-coding gene | Cytoband: 5q11.2 | ||
| Entrez ID: 202309 | HGNC ID: HGNC:26588 | Ensembl Gene: ENSG00000175857 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GAPT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GAPT | 202309 | 1552386_at | -0.0219 | 0.9813 | |
| GSE26886 | GAPT | 202309 | 1552386_at | -0.5921 | 0.0015 | |
| GSE45670 | GAPT | 202309 | 1552386_at | -0.1033 | 0.6462 | |
| GSE53622 | GAPT | 202309 | 70976 | -0.7481 | 0.0003 | |
| GSE53624 | GAPT | 202309 | 70976 | -1.4408 | 0.0000 | |
| GSE63941 | GAPT | 202309 | 1552386_at | 0.1305 | 0.3127 | |
| GSE77861 | GAPT | 202309 | 1552386_at | 0.0215 | 0.8483 | |
| GSE97050 | GAPT | 202309 | A_23_P402319 | 0.7355 | 0.2660 | |
| SRP064894 | GAPT | 202309 | RNAseq | 0.1239 | 0.7417 | |
| SRP133303 | GAPT | 202309 | RNAseq | -0.3911 | 0.1421 | |
| SRP159526 | GAPT | 202309 | RNAseq | -1.1467 | 0.1637 | |
| SRP193095 | GAPT | 202309 | RNAseq | -0.1089 | 0.6333 | |
| SRP219564 | GAPT | 202309 | RNAseq | -0.5213 | 0.3981 | |
| TCGA | GAPT | 202309 | RNAseq | -0.6415 | 0.0656 |
Upregulated datasets: 0; Downregulated datasets: 1.
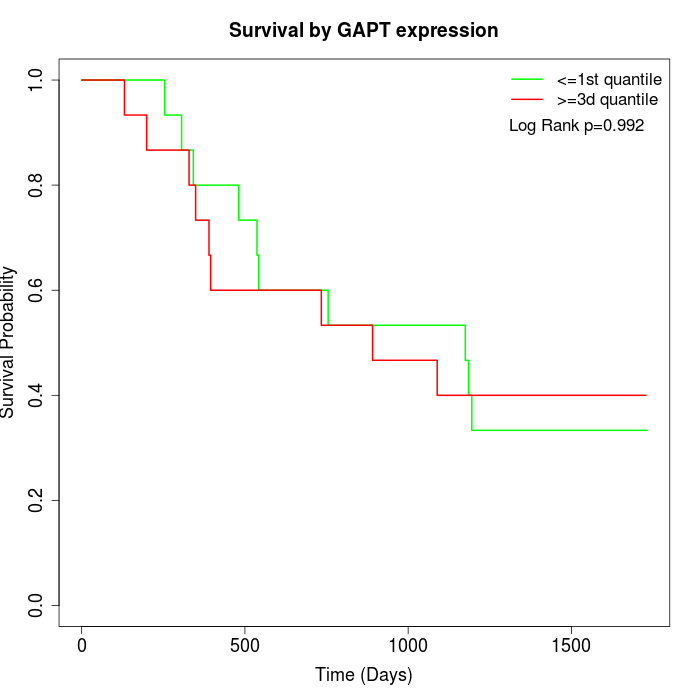
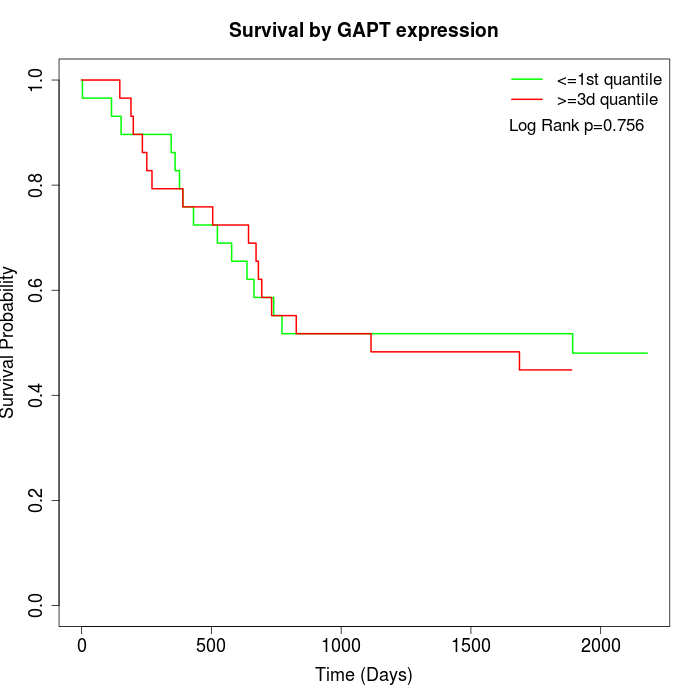
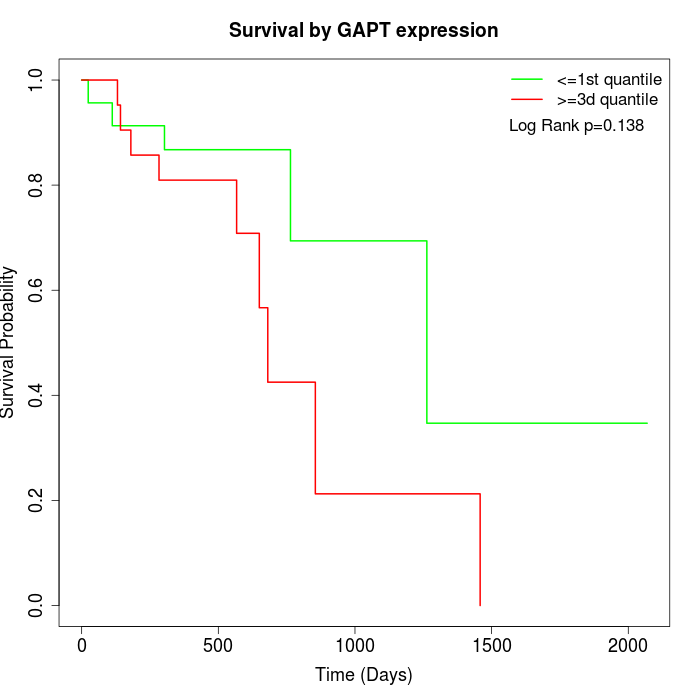
Survival by GAPT expression:
Note: Click image to view full size file.
Copy number change of GAPT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GAPT | 202309 | 0 | 13 | 17 | |
| GSE20123 | GAPT | 202309 | 0 | 13 | 17 | |
| GSE43470 | GAPT | 202309 | 1 | 12 | 30 | |
| GSE46452 | GAPT | 202309 | 0 | 28 | 31 | |
| GSE47630 | GAPT | 202309 | 2 | 16 | 22 | |
| GSE54993 | GAPT | 202309 | 8 | 1 | 61 | |
| GSE54994 | GAPT | 202309 | 2 | 19 | 32 | |
| GSE60625 | GAPT | 202309 | 0 | 0 | 11 | |
| GSE74703 | GAPT | 202309 | 1 | 9 | 26 | |
| GSE74704 | GAPT | 202309 | 0 | 6 | 14 | |
| TCGA | GAPT | 202309 | 7 | 46 | 43 |
Total number of gains: 21; Total number of losses: 163; Total Number of normals: 304.
Somatic mutations of GAPT:
Generating mutation plots.
Highly correlated genes for GAPT:
Showing top 20/350 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GAPT | CD79B | 0.817725 | 4 | 0 | 4 |
| GAPT | GIMAP5 | 0.798289 | 3 | 0 | 3 |
| GAPT | ARHGAP15 | 0.797853 | 3 | 0 | 3 |
| GAPT | CEP57 | 0.766888 | 3 | 0 | 3 |
| GAPT | CD37 | 0.764026 | 5 | 0 | 5 |
| GAPT | P2RY13 | 0.756695 | 6 | 0 | 6 |
| GAPT | CCR2 | 0.75249 | 6 | 0 | 6 |
| GAPT | NUDT21 | 0.748329 | 3 | 0 | 3 |
| GAPT | TLR10 | 0.74774 | 4 | 0 | 4 |
| GAPT | PDE4B | 0.747703 | 3 | 0 | 3 |
| GAPT | LINC01215 | 0.745246 | 5 | 0 | 4 |
| GAPT | KLRB1 | 0.743218 | 6 | 0 | 6 |
| GAPT | AOAH | 0.736658 | 4 | 0 | 4 |
| GAPT | CORO1A | 0.73268 | 6 | 0 | 5 |
| GAPT | STAP1 | 0.732326 | 5 | 0 | 5 |
| GAPT | PIK3R6 | 0.732176 | 3 | 0 | 3 |
| GAPT | PSMA5 | 0.725226 | 3 | 0 | 3 |
| GAPT | SNX20 | 0.72427 | 6 | 0 | 6 |
| GAPT | CD96 | 0.722393 | 4 | 0 | 4 |
| GAPT | INPP5D | 0.716927 | 6 | 0 | 6 |
For details and further investigation, click here