| Full name: cofilin 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 1072 | HGNC ID: HGNC:1874 | Ensembl Gene: ENSG00000172757 | OMIM ID: 601442 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CFL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04666 | Fc gamma R-mediated phagocytosis | |
| hsa04810 | Regulation of actin cytoskeleton | |
| hsa05133 | Pertussis |
Expression of CFL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFL1 | 1072 | 200021_at | 0.2762 | 0.1765 | |
| GSE20347 | CFL1 | 1072 | 200021_at | 0.2272 | 0.0400 | |
| GSE23400 | CFL1 | 1072 | 200021_at | 0.2290 | 0.0000 | |
| GSE26886 | CFL1 | 1072 | 200021_at | -0.0227 | 0.9139 | |
| GSE29001 | CFL1 | 1072 | 200021_at | 0.0889 | 0.7815 | |
| GSE38129 | CFL1 | 1072 | 200021_at | 0.2217 | 0.0058 | |
| GSE45670 | CFL1 | 1072 | 200021_at | 0.3830 | 0.0002 | |
| GSE53622 | CFL1 | 1072 | 41736 | 0.2953 | 0.0000 | |
| GSE53624 | CFL1 | 1072 | 41736 | 0.4722 | 0.0000 | |
| GSE63941 | CFL1 | 1072 | 200021_at | 0.6272 | 0.0151 | |
| GSE77861 | CFL1 | 1072 | 200021_at | 0.3599 | 0.0212 | |
| GSE97050 | CFL1 | 1072 | A_32_P148710 | 0.6452 | 0.1726 | |
| SRP007169 | CFL1 | 1072 | RNAseq | 0.6219 | 0.0489 | |
| SRP008496 | CFL1 | 1072 | RNAseq | 0.4014 | 0.0175 | |
| SRP064894 | CFL1 | 1072 | RNAseq | 0.4900 | 0.0189 | |
| SRP133303 | CFL1 | 1072 | RNAseq | 0.8472 | 0.0001 | |
| SRP159526 | CFL1 | 1072 | RNAseq | 0.1214 | 0.5416 | |
| SRP193095 | CFL1 | 1072 | RNAseq | -0.0174 | 0.9147 | |
| SRP219564 | CFL1 | 1072 | RNAseq | 0.4290 | 0.2961 | |
| TCGA | CFL1 | 1072 | RNAseq | 0.1597 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
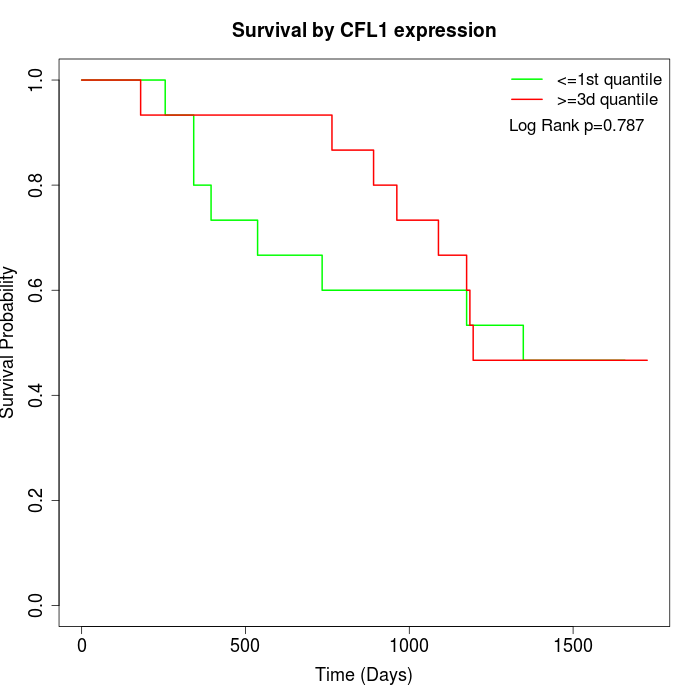
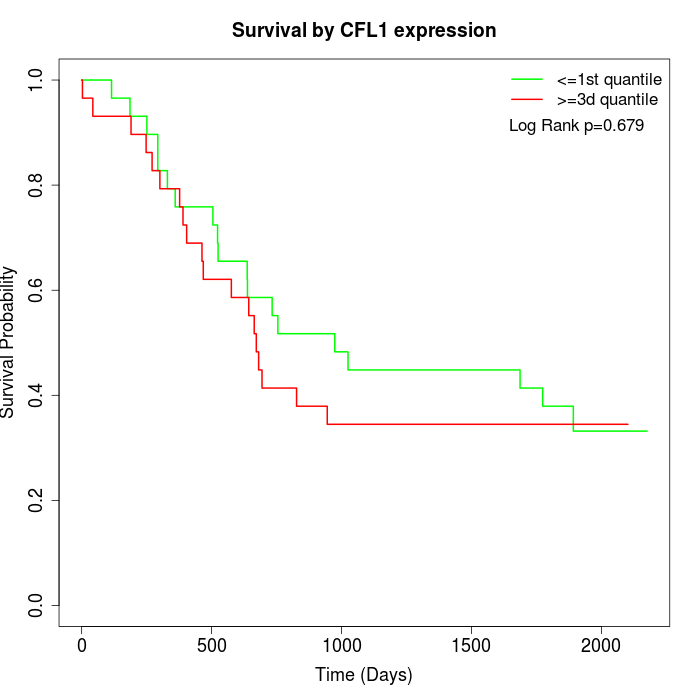
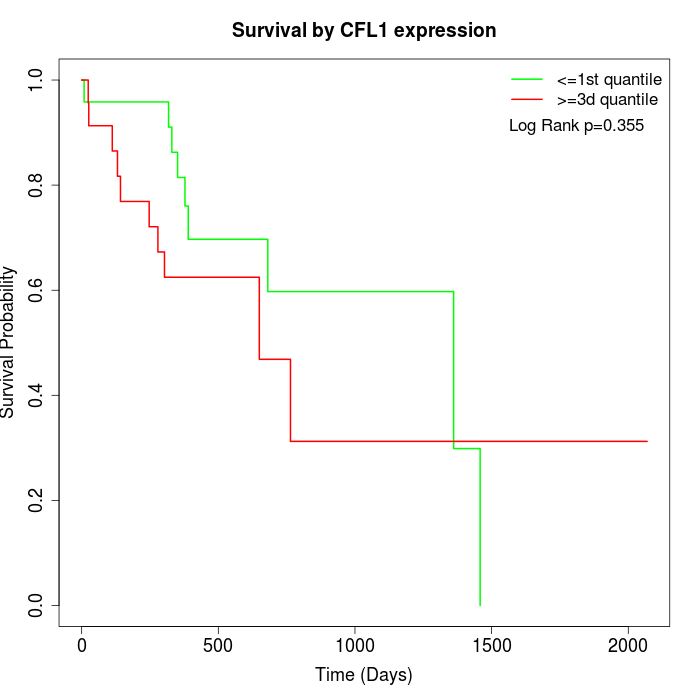
Survival by CFL1 expression:
Note: Click image to view full size file.
Copy number change of CFL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFL1 | 1072 | 8 | 5 | 17 | |
| GSE20123 | CFL1 | 1072 | 8 | 5 | 17 | |
| GSE43470 | CFL1 | 1072 | 6 | 1 | 36 | |
| GSE46452 | CFL1 | 1072 | 11 | 3 | 45 | |
| GSE47630 | CFL1 | 1072 | 7 | 5 | 28 | |
| GSE54993 | CFL1 | 1072 | 3 | 0 | 67 | |
| GSE54994 | CFL1 | 1072 | 7 | 5 | 41 | |
| GSE60625 | CFL1 | 1072 | 0 | 3 | 8 | |
| GSE74703 | CFL1 | 1072 | 4 | 0 | 32 | |
| GSE74704 | CFL1 | 1072 | 6 | 3 | 11 | |
| TCGA | CFL1 | 1072 | 23 | 7 | 66 |
Total number of gains: 83; Total number of losses: 37; Total Number of normals: 368.
Somatic mutations of CFL1:
Generating mutation plots.
Highly correlated genes for CFL1:
Showing top 20/1236 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFL1 | GRAMD1A | 0.750321 | 3 | 0 | 3 |
| CFL1 | BRMS1 | 0.740137 | 10 | 0 | 10 |
| CFL1 | TDRD5 | 0.728933 | 3 | 0 | 3 |
| CFL1 | CUL7 | 0.720577 | 3 | 0 | 3 |
| CFL1 | EME1 | 0.712259 | 5 | 0 | 5 |
| CFL1 | HSD17B1 | 0.711696 | 4 | 0 | 4 |
| CFL1 | PSMG3 | 0.70778 | 6 | 0 | 6 |
| CFL1 | ZNF670 | 0.706968 | 3 | 0 | 3 |
| CFL1 | RAN | 0.698637 | 6 | 0 | 6 |
| CFL1 | C4orf48 | 0.696247 | 5 | 0 | 5 |
| CFL1 | IRF2BPL | 0.696196 | 3 | 0 | 3 |
| CFL1 | CDCA2 | 0.696007 | 5 | 0 | 5 |
| CFL1 | NRSN2 | 0.695755 | 3 | 0 | 3 |
| CFL1 | PLEK | 0.685281 | 3 | 0 | 3 |
| CFL1 | SCYL1 | 0.68405 | 5 | 0 | 5 |
| CFL1 | OGFOD2 | 0.683503 | 5 | 0 | 5 |
| CFL1 | DPP3 | 0.683147 | 11 | 0 | 10 |
| CFL1 | RRP36 | 0.682497 | 5 | 0 | 5 |
| CFL1 | EBNA1BP2 | 0.681799 | 3 | 0 | 3 |
| CFL1 | MRPL14 | 0.679753 | 5 | 0 | 4 |
For details and further investigation, click here