| Full name: CASP8 and FADD like apoptosis regulator | Alias Symbol: CASH|Casper|CLARP|FLAME|FLIP|I-FLICE|MRIT|c-FLIP|cFLIP | ||
| Type: protein-coding gene | Cytoband: 2q33.1 | ||
| Entrez ID: 8837 | HGNC ID: HGNC:1876 | Ensembl Gene: ENSG00000003402 | OMIM ID: 603599 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CFLAR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04064 | NF-kappa B signaling pathway | |
| hsa04210 | Apoptosis | |
| hsa04668 | TNF signaling pathway | |
| hsa05142 | Chagas disease (American trypanosomiasis) |
Expression of CFLAR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFLAR | 8837 | 209939_x_at | -0.1192 | 0.8614 | |
| GSE20347 | CFLAR | 8837 | 211316_x_at | -0.3751 | 0.0712 | |
| GSE23400 | CFLAR | 8837 | 209939_x_at | -0.3821 | 0.0000 | |
| GSE26886 | CFLAR | 8837 | 209939_x_at | -1.4268 | 0.0000 | |
| GSE29001 | CFLAR | 8837 | 209939_x_at | -0.4433 | 0.1804 | |
| GSE38129 | CFLAR | 8837 | 209939_x_at | -0.1830 | 0.3251 | |
| GSE45670 | CFLAR | 8837 | 211316_x_at | -0.2057 | 0.1454 | |
| GSE53622 | CFLAR | 8837 | 65191 | 2.1301 | 0.0000 | |
| GSE53624 | CFLAR | 8837 | 65191 | 1.9007 | 0.0000 | |
| GSE63941 | CFLAR | 8837 | 209939_x_at | -0.3002 | 0.6841 | |
| GSE77861 | CFLAR | 8837 | 209939_x_at | -0.4697 | 0.1124 | |
| GSE97050 | CFLAR | 8837 | A_23_P156807 | 1.2136 | 0.0555 | |
| SRP007169 | CFLAR | 8837 | RNAseq | -2.1823 | 0.0003 | |
| SRP008496 | CFLAR | 8837 | RNAseq | -1.9842 | 0.0000 | |
| SRP064894 | CFLAR | 8837 | RNAseq | -0.8608 | 0.0000 | |
| SRP133303 | CFLAR | 8837 | RNAseq | -0.2863 | 0.0162 | |
| SRP159526 | CFLAR | 8837 | RNAseq | -0.3614 | 0.2323 | |
| SRP193095 | CFLAR | 8837 | RNAseq | -0.3754 | 0.0040 | |
| SRP219564 | CFLAR | 8837 | RNAseq | -0.2181 | 0.4234 | |
| TCGA | CFLAR | 8837 | RNAseq | -0.0644 | 0.2786 |
Upregulated datasets: 2; Downregulated datasets: 3.
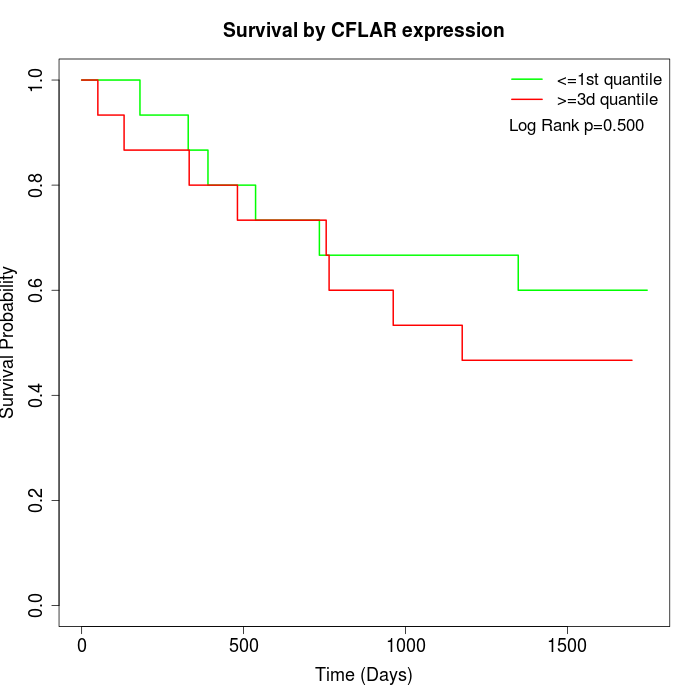
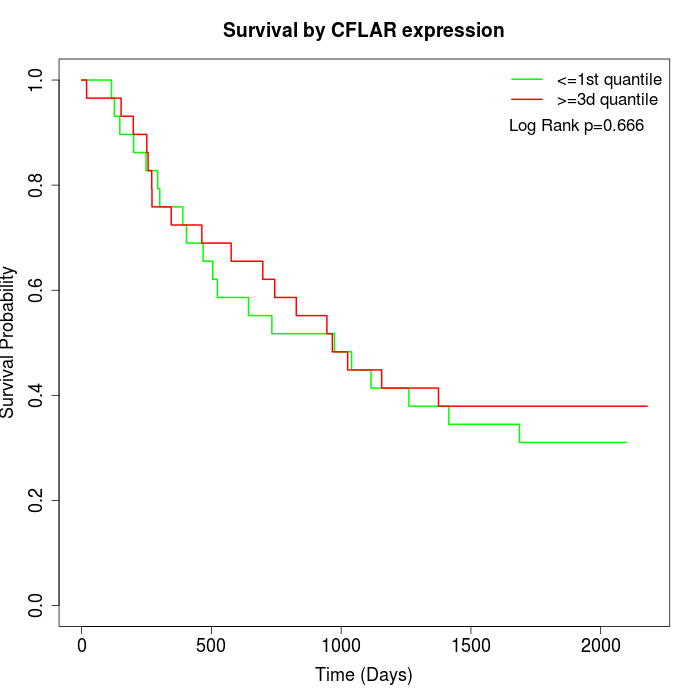
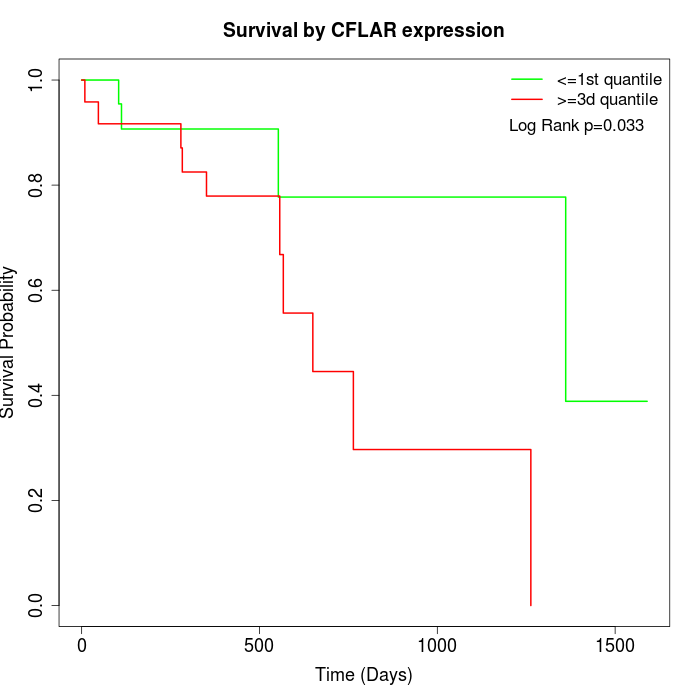
Survival by CFLAR expression:
Note: Click image to view full size file.
Copy number change of CFLAR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFLAR | 8837 | 6 | 4 | 20 | |
| GSE20123 | CFLAR | 8837 | 6 | 4 | 20 | |
| GSE43470 | CFLAR | 8837 | 1 | 1 | 41 | |
| GSE46452 | CFLAR | 8837 | 1 | 4 | 54 | |
| GSE47630 | CFLAR | 8837 | 4 | 5 | 31 | |
| GSE54993 | CFLAR | 8837 | 0 | 3 | 67 | |
| GSE54994 | CFLAR | 8837 | 10 | 8 | 35 | |
| GSE60625 | CFLAR | 8837 | 0 | 3 | 8 | |
| GSE74703 | CFLAR | 8837 | 1 | 1 | 34 | |
| GSE74704 | CFLAR | 8837 | 3 | 2 | 15 | |
| TCGA | CFLAR | 8837 | 22 | 10 | 64 |
Total number of gains: 54; Total number of losses: 45; Total Number of normals: 389.
Somatic mutations of CFLAR:
Generating mutation plots.
Highly correlated genes for CFLAR:
Showing top 20/48 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFLAR | PPIA | 0.734418 | 3 | 0 | 3 |
| CFLAR | LEPR | 0.715427 | 4 | 0 | 3 |
| CFLAR | NOMO1 | 0.715199 | 3 | 0 | 3 |
| CFLAR | SLC24A2 | 0.682016 | 3 | 0 | 3 |
| CFLAR | KIF19 | 0.679695 | 3 | 0 | 3 |
| CFLAR | TMEM26 | 0.637742 | 3 | 0 | 3 |
| CFLAR | FAM24A | 0.636097 | 4 | 0 | 3 |
| CFLAR | C19orf48 | 0.613712 | 3 | 0 | 3 |
| CFLAR | PARP14 | 0.613693 | 3 | 0 | 3 |
| CFLAR | DNAH17-AS1 | 0.613412 | 3 | 0 | 3 |
| CFLAR | BOP1 | 0.603295 | 3 | 0 | 3 |
| CFLAR | FAM222B | 0.601559 | 3 | 0 | 3 |
| CFLAR | ULBP1 | 0.599557 | 4 | 0 | 3 |
| CFLAR | P4HA2 | 0.597796 | 3 | 0 | 3 |
| CFLAR | SCARF1 | 0.594171 | 3 | 0 | 3 |
| CFLAR | CHRNA1 | 0.591193 | 4 | 0 | 3 |
| CFLAR | KRT17 | 0.590138 | 3 | 0 | 3 |
| CFLAR | LEPROT | 0.588823 | 3 | 0 | 3 |
| CFLAR | LRWD1 | 0.585596 | 3 | 0 | 3 |
| CFLAR | GBP6 | 0.583322 | 4 | 0 | 3 |
For details and further investigation, click here