| Full name: peptidylprolyl isomerase A | Alias Symbol: CYPA | ||
| Type: protein-coding gene | Cytoband: 7p13 | ||
| Entrez ID: 5478 | HGNC ID: HGNC:9253 | Ensembl Gene: ENSG00000196262 | OMIM ID: 123840 |
| Drug and gene relationship at DGIdb | |||
Expression of PPIA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | PPIA | 5478 | 91290 | 0.8492 | 0.0000 | |
| GSE53624 | PPIA | 5478 | 91290 | 0.7665 | 0.0000 | |
| GSE97050 | PPIA | 5478 | A_24_P95038 | 0.6931 | 0.0711 | |
| SRP007169 | PPIA | 5478 | RNAseq | -0.8749 | 0.0048 | |
| SRP008496 | PPIA | 5478 | RNAseq | -0.5863 | 0.0281 | |
| SRP064894 | PPIA | 5478 | RNAseq | 0.4040 | 0.1385 | |
| SRP133303 | PPIA | 5478 | RNAseq | 0.3031 | 0.0861 | |
| SRP159526 | PPIA | 5478 | RNAseq | 0.0969 | 0.7414 | |
| SRP193095 | PPIA | 5478 | RNAseq | -0.2219 | 0.2128 | |
| SRP219564 | PPIA | 5478 | RNAseq | -0.1363 | 0.6559 | |
| TCGA | PPIA | 5478 | RNAseq | 0.2920 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
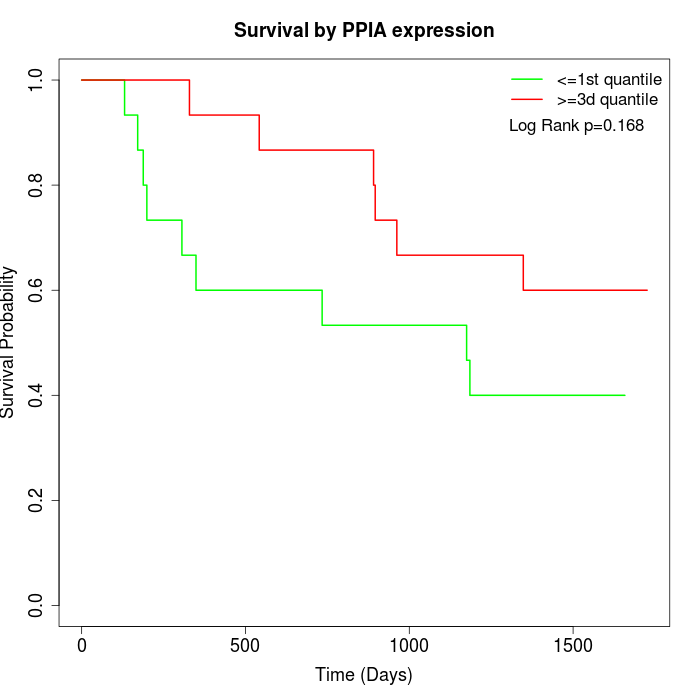
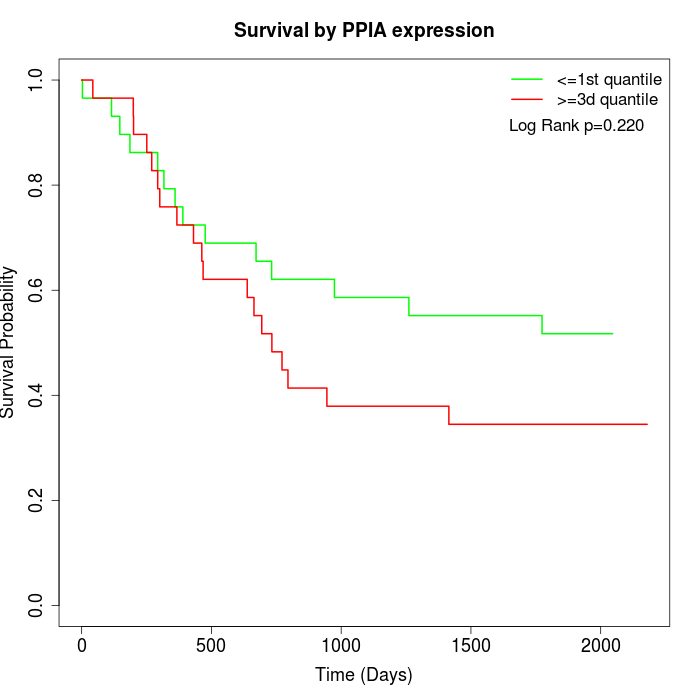
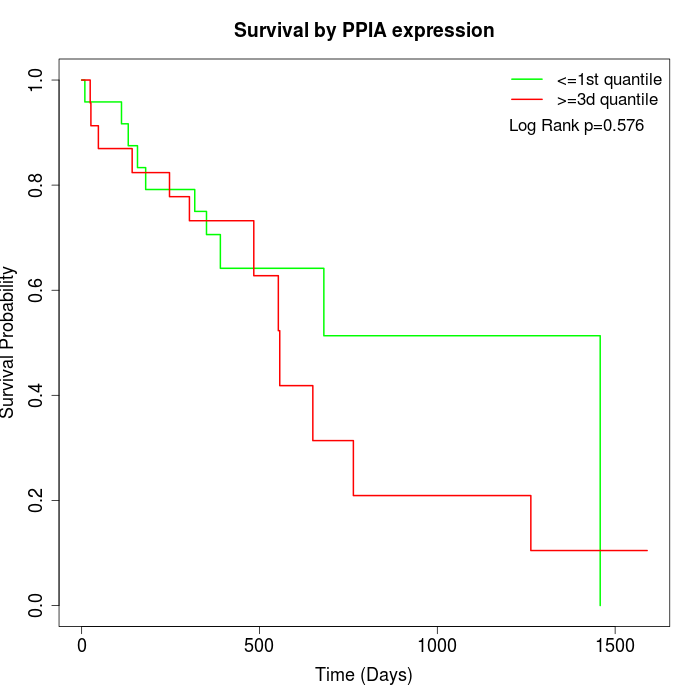
Survival by PPIA expression:
Note: Click image to view full size file.
Copy number change of PPIA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPIA | 5478 | 13 | 0 | 17 | |
| GSE20123 | PPIA | 5478 | 12 | 0 | 18 | |
| GSE43470 | PPIA | 5478 | 4 | 0 | 39 | |
| GSE46452 | PPIA | 5478 | 11 | 1 | 47 | |
| GSE47630 | PPIA | 5478 | 8 | 1 | 31 | |
| GSE54993 | PPIA | 5478 | 0 | 8 | 62 | |
| GSE54994 | PPIA | 5478 | 16 | 3 | 34 | |
| GSE60625 | PPIA | 5478 | 0 | 0 | 11 | |
| GSE74703 | PPIA | 5478 | 4 | 0 | 32 | |
| GSE74704 | PPIA | 5478 | 6 | 0 | 14 | |
| TCGA | PPIA | 5478 | 50 | 7 | 39 |
Total number of gains: 124; Total number of losses: 20; Total Number of normals: 344.
Somatic mutations of PPIA:
Generating mutation plots.
Highly correlated genes for PPIA:
Showing top 20/76 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPIA | FSCN1 | 0.743738 | 3 | 0 | 3 |
| PPIA | CFLAR | 0.734418 | 3 | 0 | 3 |
| PPIA | SLC2A1 | 0.725532 | 3 | 0 | 3 |
| PPIA | MMP11 | 0.705722 | 3 | 0 | 3 |
| PPIA | COL3A1 | 0.688841 | 3 | 0 | 3 |
| PPIA | COL10A1 | 0.685209 | 3 | 0 | 3 |
| PPIA | COL1A1 | 0.679005 | 3 | 0 | 3 |
| PPIA | TP63 | 0.678222 | 3 | 0 | 3 |
| PPIA | LAMC2 | 0.676373 | 3 | 0 | 3 |
| PPIA | CBX3 | 0.670293 | 3 | 0 | 3 |
| PPIA | DLGAP5 | 0.665427 | 3 | 0 | 3 |
| PPIA | SPP1 | 0.664505 | 3 | 0 | 3 |
| PPIA | KRT17 | 0.662968 | 3 | 0 | 3 |
| PPIA | CDCA8 | 0.652871 | 3 | 0 | 3 |
| PPIA | POSTN | 0.648622 | 3 | 0 | 3 |
| PPIA | ECT2 | 0.646713 | 3 | 0 | 3 |
| PPIA | EXO1 | 0.646413 | 3 | 0 | 3 |
| PPIA | EXT1 | 0.646034 | 3 | 0 | 3 |
| PPIA | GPRIN1 | 0.645587 | 3 | 0 | 3 |
| PPIA | TSPO2 | 0.644579 | 3 | 0 | 3 |
For details and further investigation, click here