| Full name: prolyl 4-hydroxylase subunit alpha 2 | Alias Symbol: C-P4Halpha(II) | ||
| Type: protein-coding gene | Cytoband: 5q31.1 | ||
| Entrez ID: 8974 | HGNC ID: HGNC:8547 | Ensembl Gene: ENSG00000072682 | OMIM ID: 600608 |
| Drug and gene relationship at DGIdb | |||
Expression of P4HA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | P4HA2 | 8974 | 57694 | 0.8261 | 0.0000 | |
| GSE53624 | P4HA2 | 8974 | 57694 | 0.8796 | 0.0000 | |
| GSE97050 | P4HA2 | 8974 | A_33_P3394933 | 0.3600 | 0.2084 | |
| SRP007169 | P4HA2 | 8974 | RNAseq | 0.9370 | 0.0577 | |
| SRP008496 | P4HA2 | 8974 | RNAseq | 0.7928 | 0.0045 | |
| SRP064894 | P4HA2 | 8974 | RNAseq | 1.4440 | 0.0000 | |
| SRP133303 | P4HA2 | 8974 | RNAseq | 1.8550 | 0.0000 | |
| SRP159526 | P4HA2 | 8974 | RNAseq | 0.7521 | 0.0313 | |
| SRP193095 | P4HA2 | 8974 | RNAseq | 1.1811 | 0.0000 | |
| SRP219564 | P4HA2 | 8974 | RNAseq | 1.4679 | 0.0095 | |
| TCGA | P4HA2 | 8974 | RNAseq | 0.1057 | 0.2288 |
Upregulated datasets: 4; Downregulated datasets: 0.
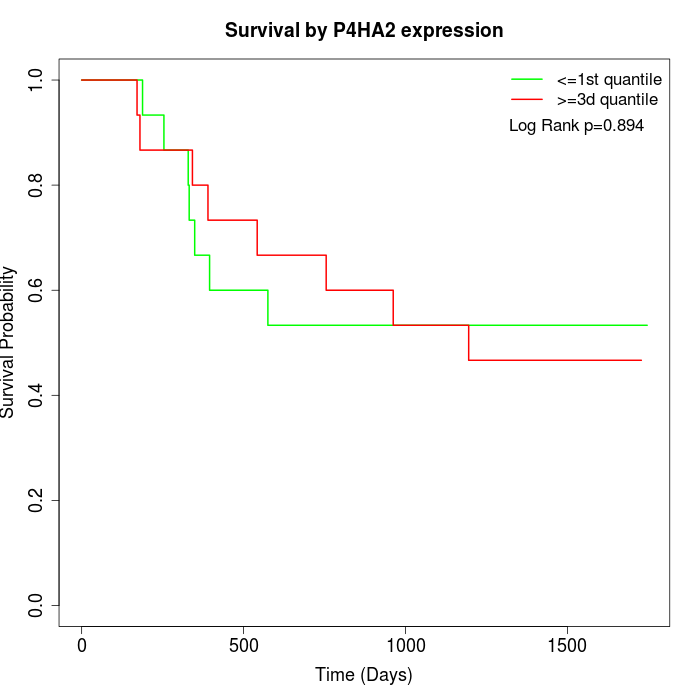
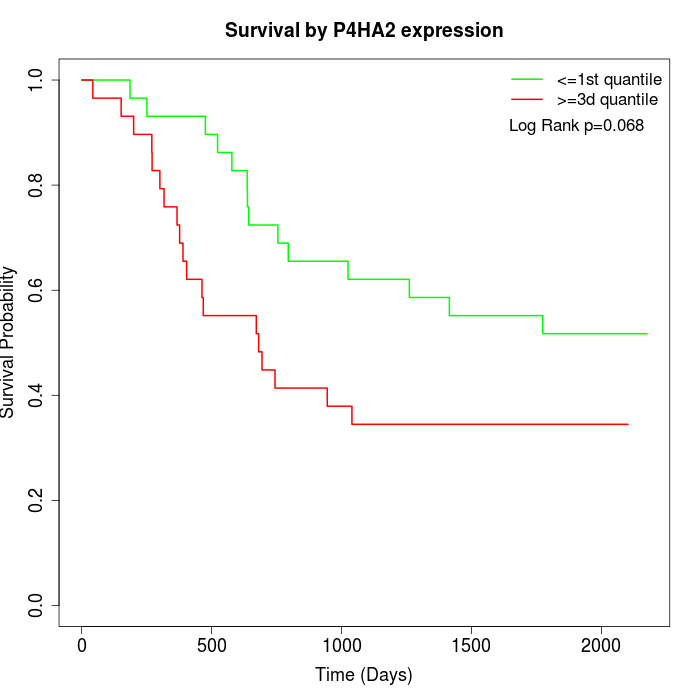
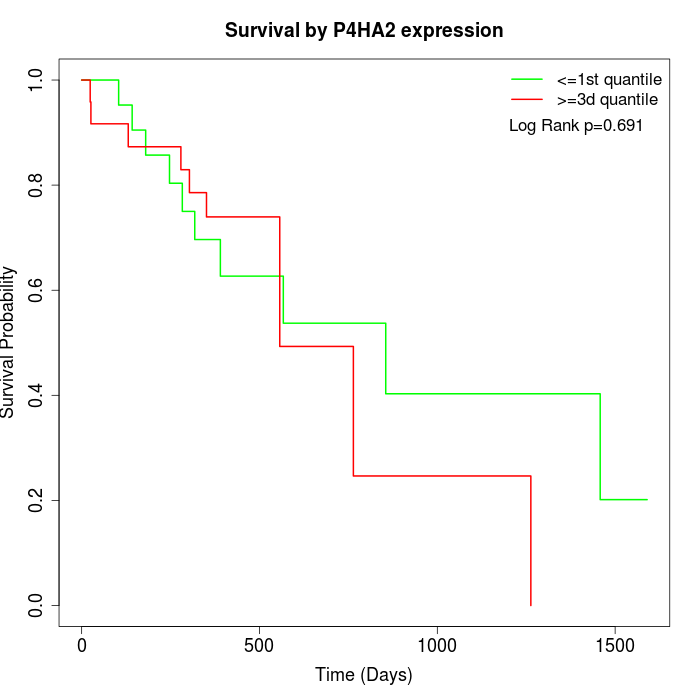
Survival by P4HA2 expression:
Note: Click image to view full size file.
Copy number change of P4HA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | P4HA2 | 8974 | 1 | 11 | 18 | |
| GSE20123 | P4HA2 | 8974 | 1 | 11 | 18 | |
| GSE43470 | P4HA2 | 8974 | 2 | 9 | 32 | |
| GSE46452 | P4HA2 | 8974 | 0 | 27 | 32 | |
| GSE47630 | P4HA2 | 8974 | 0 | 21 | 19 | |
| GSE54993 | P4HA2 | 8974 | 9 | 1 | 60 | |
| GSE54994 | P4HA2 | 8974 | 2 | 14 | 37 | |
| GSE60625 | P4HA2 | 8974 | 0 | 0 | 11 | |
| GSE74703 | P4HA2 | 8974 | 2 | 6 | 28 | |
| GSE74704 | P4HA2 | 8974 | 1 | 5 | 14 | |
| TCGA | P4HA2 | 8974 | 3 | 39 | 54 |
Total number of gains: 21; Total number of losses: 144; Total Number of normals: 323.
Somatic mutations of P4HA2:
Generating mutation plots.
Highly correlated genes for P4HA2:
Showing top 20/121 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| P4HA2 | RCN3 | 0.751293 | 3 | 0 | 3 |
| P4HA2 | KIF26B | 0.747255 | 3 | 0 | 3 |
| P4HA2 | CHPF2 | 0.741589 | 3 | 0 | 3 |
| P4HA2 | TSPO2 | 0.74117 | 3 | 0 | 3 |
| P4HA2 | FADS3 | 0.735601 | 3 | 0 | 3 |
| P4HA2 | IL15RA | 0.72491 | 3 | 0 | 3 |
| P4HA2 | MFSD10 | 0.716119 | 3 | 0 | 3 |
| P4HA2 | BGN | 0.715253 | 3 | 0 | 3 |
| P4HA2 | MMP11 | 0.704158 | 3 | 0 | 3 |
| P4HA2 | SH2D2A | 0.702567 | 3 | 0 | 3 |
| P4HA2 | SPP1 | 0.702048 | 3 | 0 | 3 |
| P4HA2 | EIF3B | 0.700822 | 3 | 0 | 3 |
| P4HA2 | C1QTNF6 | 0.693227 | 3 | 0 | 3 |
| P4HA2 | COL10A1 | 0.689222 | 3 | 0 | 3 |
| P4HA2 | TGFBI | 0.682296 | 3 | 0 | 3 |
| P4HA2 | PML | 0.681619 | 3 | 0 | 3 |
| P4HA2 | GALNS | 0.681154 | 3 | 0 | 3 |
| P4HA2 | ADAMTS6 | 0.681002 | 3 | 0 | 3 |
| P4HA2 | COL27A1 | 0.675553 | 4 | 0 | 3 |
| P4HA2 | EPHB2 | 0.674796 | 3 | 0 | 3 |
For details and further investigation, click here