| Full name: CF transmembrane conductance regulator | Alias Symbol: MRP7|ABC35|TNR-CFTR|dJ760C5.1|CFTR/MRP | ||
| Type: protein-coding gene | Cytoband: 7q31.2 | ||
| Entrez ID: 1080 | HGNC ID: HGNC:1884 | Ensembl Gene: ENSG00000001626 | OMIM ID: 602421 |
| Drug and gene relationship at DGIdb | |||
CFTR involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway | |
| hsa04152 | AMPK signaling pathway | |
| hsa04972 | Pancreatic secretion | |
| hsa04976 | Bile secretion | |
| hsa05110 | Vibrio cholerae infection |
Expression of CFTR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CFTR | 1080 | 205043_at | -0.2846 | 0.4570 | |
| GSE20347 | CFTR | 1080 | 205043_at | -0.3044 | 0.0011 | |
| GSE23400 | CFTR | 1080 | 217026_at | -0.0434 | 0.1941 | |
| GSE26886 | CFTR | 1080 | 205043_at | -0.1016 | 0.8916 | |
| GSE29001 | CFTR | 1080 | 205043_at | -1.2069 | 0.0000 | |
| GSE38129 | CFTR | 1080 | 205043_at | -0.4046 | 0.0007 | |
| GSE45670 | CFTR | 1080 | 205043_at | -0.1957 | 0.4914 | |
| GSE53622 | CFTR | 1080 | 79420 | -1.6586 | 0.0000 | |
| GSE53624 | CFTR | 1080 | 79420 | -3.2351 | 0.0000 | |
| GSE63941 | CFTR | 1080 | 205043_at | -1.6218 | 0.0017 | |
| GSE77861 | CFTR | 1080 | 205043_at | -0.0312 | 0.8603 | |
| SRP064894 | CFTR | 1080 | RNAseq | -0.8978 | 0.2049 | |
| SRP133303 | CFTR | 1080 | RNAseq | -0.1548 | 0.8655 | |
| SRP159526 | CFTR | 1080 | RNAseq | -1.7258 | 0.0079 | |
| TCGA | CFTR | 1080 | RNAseq | -1.1913 | 0.0699 |
Upregulated datasets: 0; Downregulated datasets: 5.
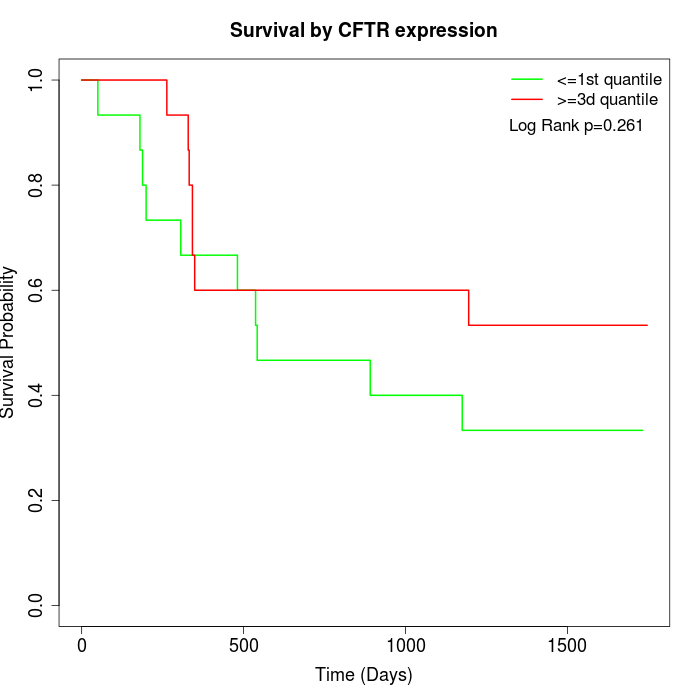
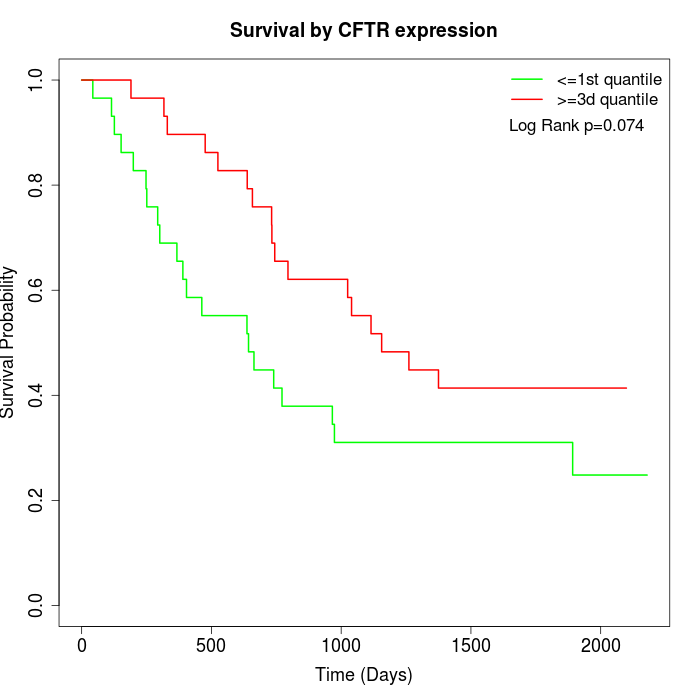
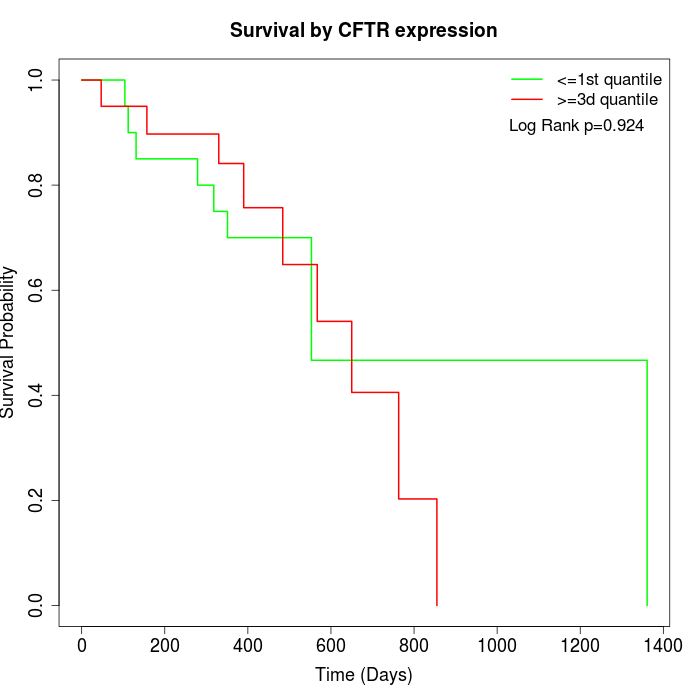
Survival by CFTR expression:
Note: Click image to view full size file.
Copy number change of CFTR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CFTR | 1080 | 8 | 2 | 20 | |
| GSE20123 | CFTR | 1080 | 9 | 2 | 19 | |
| GSE43470 | CFTR | 1080 | 4 | 2 | 37 | |
| GSE46452 | CFTR | 1080 | 8 | 1 | 50 | |
| GSE47630 | CFTR | 1080 | 7 | 4 | 29 | |
| GSE54993 | CFTR | 1080 | 2 | 7 | 61 | |
| GSE54994 | CFTR | 1080 | 12 | 5 | 36 | |
| GSE60625 | CFTR | 1080 | 0 | 0 | 11 | |
| GSE74703 | CFTR | 1080 | 4 | 2 | 30 | |
| GSE74704 | CFTR | 1080 | 6 | 2 | 12 | |
| TCGA | CFTR | 1080 | 33 | 17 | 46 |
Total number of gains: 93; Total number of losses: 44; Total Number of normals: 351.
Somatic mutations of CFTR:
Generating mutation plots.
Highly correlated genes for CFTR:
Showing top 20/631 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CFTR | SLITRK4 | 0.715123 | 3 | 0 | 3 |
| CFTR | SCNN1B | 0.681303 | 7 | 0 | 7 |
| CFTR | CYP2C9 | 0.680155 | 7 | 0 | 6 |
| CFTR | CEACAM5 | 0.670091 | 7 | 0 | 6 |
| CFTR | SCUBE3 | 0.668912 | 3 | 0 | 3 |
| CFTR | YOD1 | 0.666663 | 6 | 0 | 5 |
| CFTR | CTTNBP2 | 0.662633 | 5 | 0 | 5 |
| CFTR | MID2 | 0.662578 | 4 | 0 | 4 |
| CFTR | BPIFB1 | 0.662188 | 5 | 0 | 4 |
| CFTR | PF4 | 0.661946 | 3 | 0 | 3 |
| CFTR | ATP10B | 0.660511 | 7 | 0 | 6 |
| CFTR | CYP2J2 | 0.658328 | 6 | 0 | 5 |
| CFTR | GDPD3 | 0.657285 | 7 | 0 | 5 |
| CFTR | CWH43 | 0.656106 | 5 | 0 | 4 |
| CFTR | CXCR5 | 0.653609 | 3 | 0 | 3 |
| CFTR | CHST9 | 0.652973 | 4 | 0 | 4 |
| CFTR | DHRS9 | 0.652004 | 5 | 0 | 5 |
| CFTR | GOLPH3L | 0.65164 | 6 | 0 | 6 |
| CFTR | CYP2C18 | 0.650333 | 7 | 0 | 6 |
| CFTR | PCDH1 | 0.649927 | 6 | 0 | 6 |
For details and further investigation, click here