| Full name: carbohydrate sulfotransferase 9 | Alias Symbol: GALNAC4ST-2|GALNAC-4-ST2 | ||
| Type: protein-coding gene | Cytoband: 18q11.2 | ||
| Entrez ID: 83539 | HGNC ID: HGNC:19898 | Ensembl Gene: ENSG00000154080 | OMIM ID: 610191 |
| Drug and gene relationship at DGIdb | |||
Expression of CHST9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHST9 | 83539 | 223737_x_at | -0.7426 | 0.6938 | |
| GSE26886 | CHST9 | 83539 | 223737_x_at | -0.3483 | 0.4474 | |
| GSE45670 | CHST9 | 83539 | 223737_x_at | -0.8879 | 0.0745 | |
| GSE53622 | CHST9 | 83539 | 10877 | -3.2027 | 0.0000 | |
| GSE53624 | CHST9 | 83539 | 10877 | -3.1972 | 0.0000 | |
| GSE63941 | CHST9 | 83539 | 223737_x_at | 0.2894 | 0.6546 | |
| GSE77861 | CHST9 | 83539 | 223737_x_at | 0.0466 | 0.8978 | |
| SRP064894 | CHST9 | 83539 | RNAseq | -2.4691 | 0.0000 | |
| SRP133303 | CHST9 | 83539 | RNAseq | -0.7159 | 0.4262 | |
| SRP159526 | CHST9 | 83539 | RNAseq | -2.1418 | 0.0050 | |
| SRP193095 | CHST9 | 83539 | RNAseq | 0.5153 | 0.1473 | |
| SRP219564 | CHST9 | 83539 | RNAseq | -0.6332 | 0.3674 | |
| TCGA | CHST9 | 83539 | RNAseq | -0.8671 | 0.1793 |
Upregulated datasets: 0; Downregulated datasets: 4.
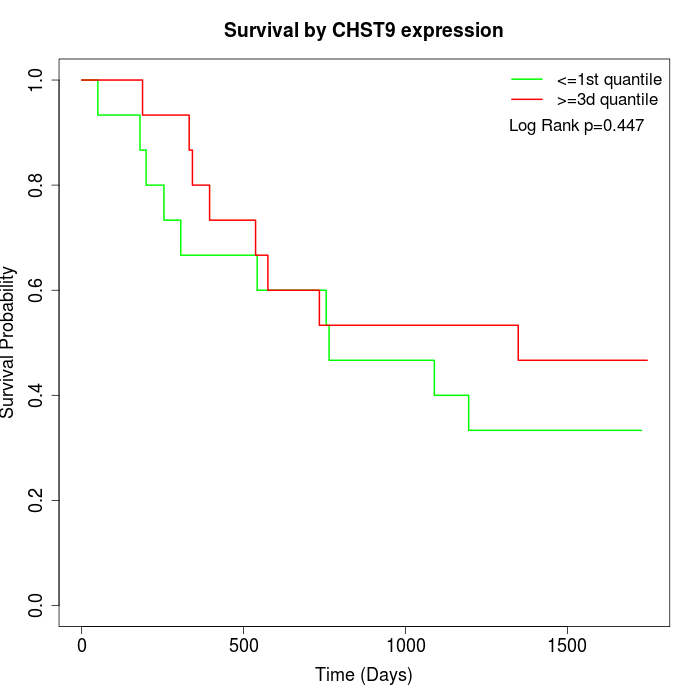
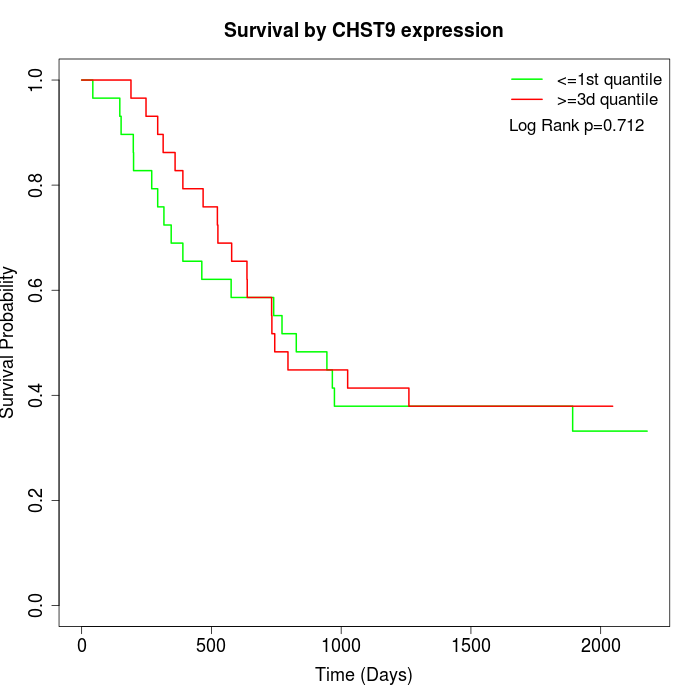
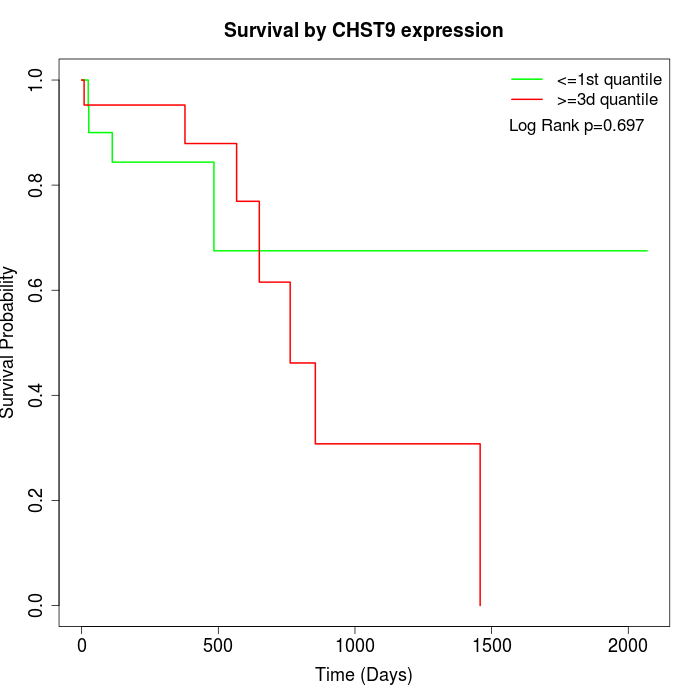
Survival by CHST9 expression:
Note: Click image to view full size file.
Copy number change of CHST9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHST9 | 83539 | 2 | 5 | 23 | |
| GSE20123 | CHST9 | 83539 | 2 | 5 | 23 | |
| GSE43470 | CHST9 | 83539 | 0 | 2 | 41 | |
| GSE46452 | CHST9 | 83539 | 1 | 25 | 33 | |
| GSE47630 | CHST9 | 83539 | 5 | 19 | 16 | |
| GSE54993 | CHST9 | 83539 | 7 | 1 | 62 | |
| GSE54994 | CHST9 | 83539 | 3 | 14 | 36 | |
| GSE60625 | CHST9 | 83539 | 0 | 4 | 7 | |
| GSE74703 | CHST9 | 83539 | 0 | 2 | 34 | |
| GSE74704 | CHST9 | 83539 | 1 | 4 | 15 | |
| TCGA | CHST9 | 83539 | 16 | 33 | 47 |
Total number of gains: 37; Total number of losses: 114; Total Number of normals: 337.
Somatic mutations of CHST9:
Generating mutation plots.
Highly correlated genes for CHST9:
Showing top 20/180 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHST9 | ACADL | 0.697123 | 5 | 0 | 5 |
| CHST9 | ADH1C | 0.67865 | 5 | 0 | 5 |
| CHST9 | COX7B2 | 0.674793 | 3 | 0 | 3 |
| CHST9 | ARL1 | 0.664419 | 3 | 0 | 3 |
| CHST9 | B3GALT5 | 0.663045 | 4 | 0 | 4 |
| CHST9 | BEX1 | 0.653262 | 3 | 0 | 3 |
| CHST9 | CFTR | 0.652973 | 4 | 0 | 4 |
| CHST9 | TOX3 | 0.652288 | 4 | 0 | 3 |
| CHST9 | IYD | 0.651949 | 3 | 0 | 3 |
| CHST9 | CHAD | 0.649289 | 3 | 0 | 3 |
| CHST9 | ERBB4 | 0.643159 | 3 | 0 | 3 |
| CHST9 | CRYM | 0.64233 | 3 | 0 | 3 |
| CHST9 | HMGCS2 | 0.639917 | 5 | 0 | 4 |
| CHST9 | FAM3B | 0.634331 | 5 | 0 | 4 |
| CHST9 | SP1 | 0.632992 | 4 | 0 | 3 |
| CHST9 | TMPRSS2 | 0.627351 | 4 | 0 | 4 |
| CHST9 | SNX31 | 0.619702 | 5 | 0 | 5 |
| CHST9 | TACC1 | 0.618745 | 4 | 0 | 4 |
| CHST9 | GLYATL2 | 0.615353 | 3 | 0 | 3 |
| CHST9 | BPIFB1 | 0.615062 | 5 | 0 | 4 |
For details and further investigation, click here