| Full name: CHM Rab escort protein | Alias Symbol: REP-1 | ||
| Type: protein-coding gene | Cytoband: Xq21.2 | ||
| Entrez ID: 1121 | HGNC ID: HGNC:1940 | Ensembl Gene: ENSG00000188419 | OMIM ID: 300390 |
| Drug and gene relationship at DGIdb | |||
Expression of CHM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHM | 1121 | 227871_at | -0.1586 | 0.6292 | |
| GSE20347 | CHM | 1121 | 207099_s_at | 0.0197 | 0.8288 | |
| GSE23400 | CHM | 1121 | 207099_s_at | 0.0235 | 0.4166 | |
| GSE26886 | CHM | 1121 | 227871_at | -0.7696 | 0.0023 | |
| GSE29001 | CHM | 1121 | 207099_s_at | -0.0538 | 0.6845 | |
| GSE38129 | CHM | 1121 | 207099_s_at | 0.0303 | 0.7041 | |
| GSE45670 | CHM | 1121 | 227871_at | -0.0026 | 0.9912 | |
| GSE53622 | CHM | 1121 | 33159 | 0.0443 | 0.5794 | |
| GSE53624 | CHM | 1121 | 33159 | 0.0476 | 0.6671 | |
| GSE63941 | CHM | 1121 | 227871_at | -0.8225 | 0.4146 | |
| GSE77861 | CHM | 1121 | 227871_at | -0.0403 | 0.8654 | |
| GSE97050 | CHM | 1121 | A_23_P22548 | 0.0317 | 0.8819 | |
| SRP007169 | CHM | 1121 | RNAseq | -0.0737 | 0.8474 | |
| SRP008496 | CHM | 1121 | RNAseq | 0.2973 | 0.2345 | |
| SRP064894 | CHM | 1121 | RNAseq | 0.1808 | 0.4858 | |
| SRP133303 | CHM | 1121 | RNAseq | 0.2708 | 0.1664 | |
| SRP159526 | CHM | 1121 | RNAseq | -0.0492 | 0.8306 | |
| SRP193095 | CHM | 1121 | RNAseq | 0.0402 | 0.6469 | |
| SRP219564 | CHM | 1121 | RNAseq | -0.0220 | 0.9508 | |
| TCGA | CHM | 1121 | RNAseq | -0.0949 | 0.1102 |
Upregulated datasets: 0; Downregulated datasets: 0.
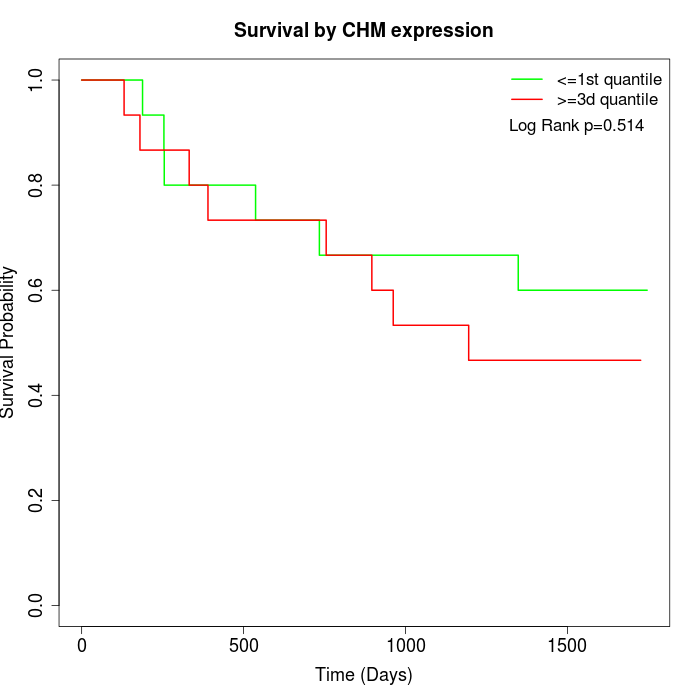
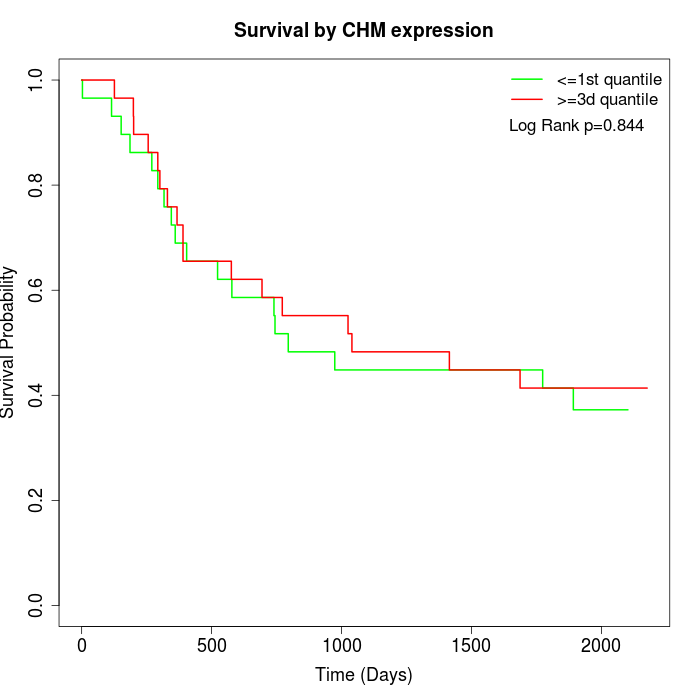
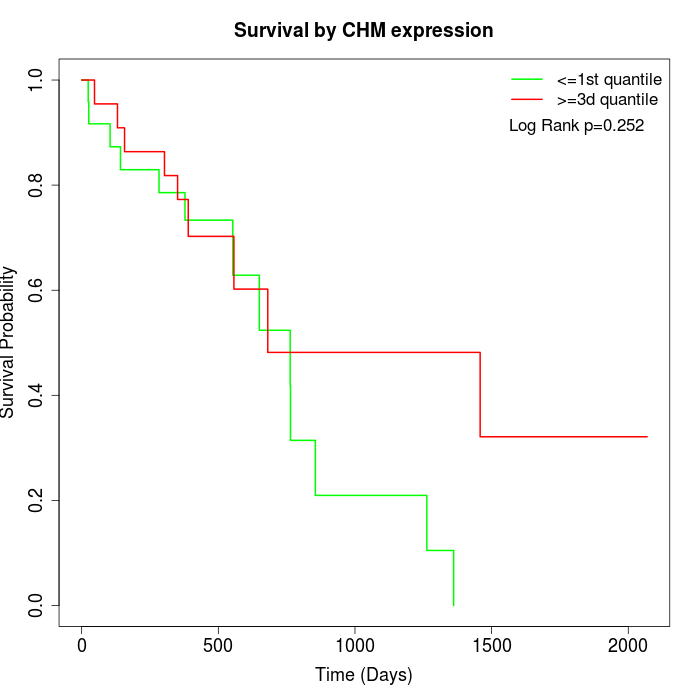
Survival by CHM expression:
Note: Click image to view full size file.
Copy number change of CHM:
No record found for this gene.
Somatic mutations of CHM:
Generating mutation plots.
Highly correlated genes for CHM:
Showing top 20/336 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHM | UHRF1BP1L | 0.828273 | 4 | 0 | 4 |
| CHM | TYW3 | 0.818398 | 3 | 0 | 3 |
| CHM | DYNC1LI2 | 0.804003 | 3 | 0 | 3 |
| CHM | INTS12 | 0.786552 | 3 | 0 | 3 |
| CHM | ARRDC3 | 0.761002 | 3 | 0 | 3 |
| CHM | PPM1D | 0.749947 | 4 | 0 | 4 |
| CHM | TM2D3 | 0.747762 | 3 | 0 | 3 |
| CHM | RHOT1 | 0.742549 | 3 | 0 | 3 |
| CHM | ZNF717 | 0.742161 | 3 | 0 | 3 |
| CHM | MAP4K5 | 0.735991 | 3 | 0 | 3 |
| CHM | C12orf29 | 0.727881 | 3 | 0 | 3 |
| CHM | HMGN5 | 0.727386 | 4 | 0 | 4 |
| CHM | MYSM1 | 0.727093 | 3 | 0 | 3 |
| CHM | ALG9 | 0.72573 | 3 | 0 | 3 |
| CHM | OR7E24 | 0.725518 | 3 | 0 | 3 |
| CHM | PAPSS2 | 0.724872 | 3 | 0 | 3 |
| CHM | NHS | 0.724478 | 3 | 0 | 3 |
| CHM | KIAA1191 | 0.723932 | 3 | 0 | 3 |
| CHM | COPS4 | 0.713686 | 4 | 0 | 3 |
| CHM | SNUPN | 0.711969 | 3 | 0 | 3 |
For details and further investigation, click here