| Full name: protein phosphatase, Mg2+/Mn2+ dependent 1D | Alias Symbol: Wip1|PP2C-DELTA | ||
| Type: protein-coding gene | Cytoband: 17q23.3 | ||
| Entrez ID: 8493 | HGNC ID: HGNC:9277 | Ensembl Gene: ENSG00000170836 | OMIM ID: 605100 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PPM1D involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04115 | p53 signaling pathway |
Expression of PPM1D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PPM1D | 8493 | 204566_at | -0.4772 | 0.3287 | |
| GSE20347 | PPM1D | 8493 | 204566_at | -0.5091 | 0.0233 | |
| GSE23400 | PPM1D | 8493 | 204566_at | -0.1884 | 0.0003 | |
| GSE26886 | PPM1D | 8493 | 204566_at | -1.1841 | 0.0004 | |
| GSE29001 | PPM1D | 8493 | 204566_at | -0.5061 | 0.1037 | |
| GSE38129 | PPM1D | 8493 | 204566_at | -0.3736 | 0.0363 | |
| GSE45670 | PPM1D | 8493 | 204566_at | -0.3055 | 0.0605 | |
| GSE53622 | PPM1D | 8493 | 5988 | -0.2274 | 0.0000 | |
| GSE53624 | PPM1D | 8493 | 5988 | -0.2853 | 0.0000 | |
| GSE63941 | PPM1D | 8493 | 204566_at | -0.8341 | 0.0555 | |
| GSE77861 | PPM1D | 8493 | 230330_at | 0.1523 | 0.2780 | |
| GSE97050 | PPM1D | 8493 | A_33_P3302696 | -0.0335 | 0.8881 | |
| SRP007169 | PPM1D | 8493 | RNAseq | -1.0460 | 0.0042 | |
| SRP008496 | PPM1D | 8493 | RNAseq | -0.8745 | 0.0000 | |
| SRP064894 | PPM1D | 8493 | RNAseq | -0.5369 | 0.0023 | |
| SRP133303 | PPM1D | 8493 | RNAseq | 0.0603 | 0.6030 | |
| SRP159526 | PPM1D | 8493 | RNAseq | 0.0855 | 0.6375 | |
| SRP193095 | PPM1D | 8493 | RNAseq | -0.3746 | 0.0000 | |
| SRP219564 | PPM1D | 8493 | RNAseq | -0.5076 | 0.0472 | |
| TCGA | PPM1D | 8493 | RNAseq | -0.0866 | 0.1451 |
Upregulated datasets: 0; Downregulated datasets: 2.
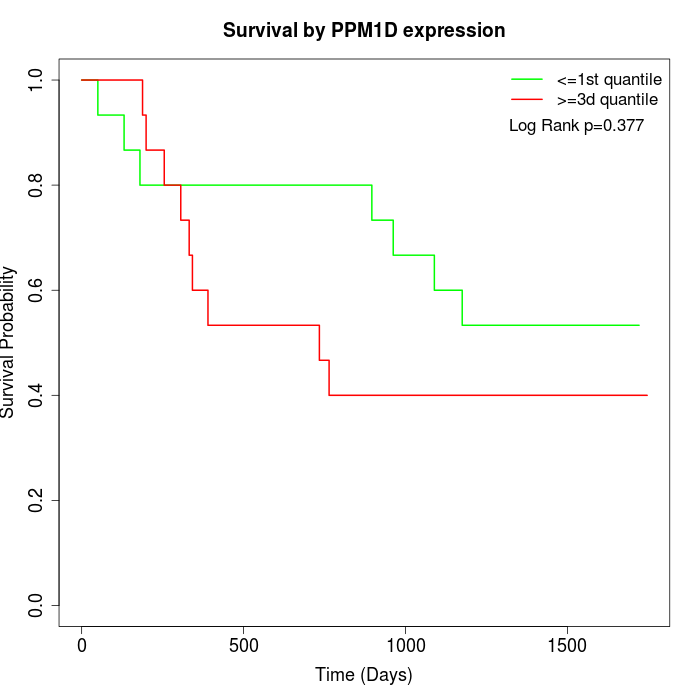
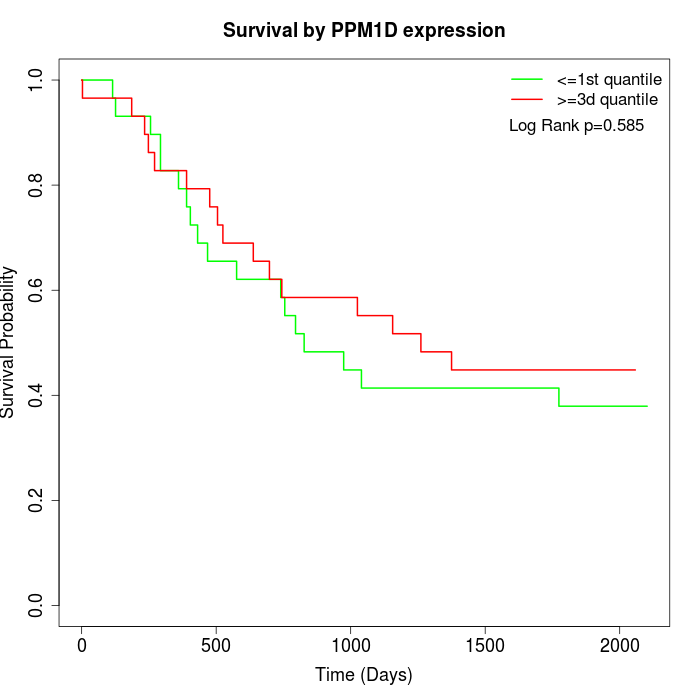
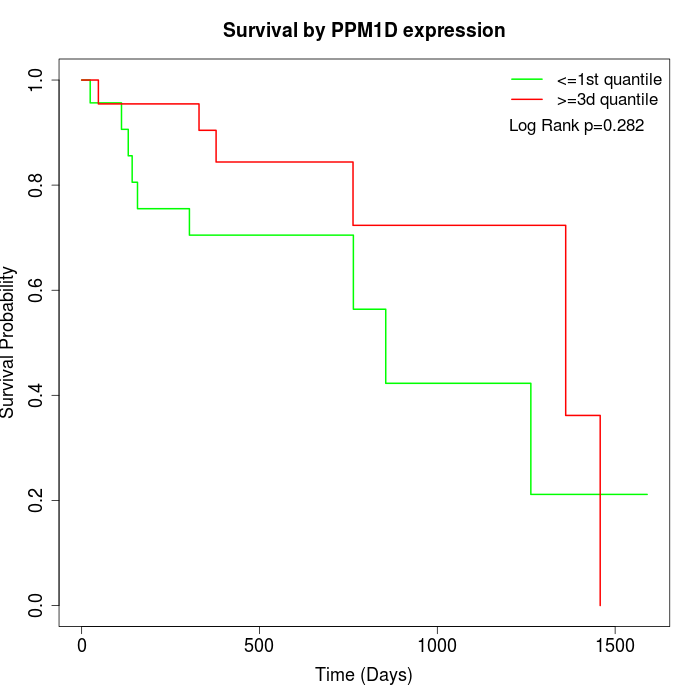
Survival by PPM1D expression:
Note: Click image to view full size file.
Copy number change of PPM1D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PPM1D | 8493 | 5 | 1 | 24 | |
| GSE20123 | PPM1D | 8493 | 5 | 1 | 24 | |
| GSE43470 | PPM1D | 8493 | 5 | 0 | 38 | |
| GSE46452 | PPM1D | 8493 | 32 | 0 | 27 | |
| GSE47630 | PPM1D | 8493 | 7 | 1 | 32 | |
| GSE54993 | PPM1D | 8493 | 2 | 5 | 63 | |
| GSE54994 | PPM1D | 8493 | 9 | 5 | 39 | |
| GSE60625 | PPM1D | 8493 | 4 | 0 | 7 | |
| GSE74703 | PPM1D | 8493 | 5 | 0 | 31 | |
| GSE74704 | PPM1D | 8493 | 4 | 1 | 15 | |
| TCGA | PPM1D | 8493 | 29 | 7 | 60 |
Total number of gains: 107; Total number of losses: 21; Total Number of normals: 360.
Somatic mutations of PPM1D:
Generating mutation plots.
Highly correlated genes for PPM1D:
Showing top 20/420 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PPM1D | SS18 | 0.792622 | 3 | 0 | 3 |
| PPM1D | CASP3 | 0.790585 | 3 | 0 | 3 |
| PPM1D | UBQLN1 | 0.768619 | 3 | 0 | 3 |
| PPM1D | ENDOD1 | 0.76772 | 3 | 0 | 3 |
| PPM1D | USP32 | 0.762232 | 3 | 0 | 3 |
| PPM1D | CCNDBP1 | 0.757204 | 4 | 0 | 4 |
| PPM1D | CHM | 0.749947 | 4 | 0 | 4 |
| PPM1D | SPTLC2 | 0.747205 | 3 | 0 | 3 |
| PPM1D | ZNF514 | 0.744378 | 3 | 0 | 3 |
| PPM1D | FNIP2 | 0.74262 | 3 | 0 | 3 |
| PPM1D | HSPA4 | 0.735389 | 3 | 0 | 3 |
| PPM1D | DOK6 | 0.730868 | 4 | 0 | 3 |
| PPM1D | ARHGEF7 | 0.716944 | 3 | 0 | 3 |
| PPM1D | CUL1 | 0.710964 | 3 | 0 | 3 |
| PPM1D | EIF2S2 | 0.708402 | 3 | 0 | 3 |
| PPM1D | C16orf72 | 0.708049 | 4 | 0 | 3 |
| PPM1D | ADAMTS9 | 0.70619 | 3 | 0 | 3 |
| PPM1D | TUBB4B | 0.705745 | 3 | 0 | 3 |
| PPM1D | PKNOX2 | 0.694745 | 3 | 0 | 3 |
| PPM1D | PTGR1 | 0.690409 | 4 | 0 | 3 |
For details and further investigation, click here