| Full name: claudin 10 | Alias Symbol: OSP-L|CPETRL3 | ||
| Type: protein-coding gene | Cytoband: 13q32.1 | ||
| Entrez ID: 9071 | HGNC ID: HGNC:2033 | Ensembl Gene: ENSG00000134873 | OMIM ID: 617579 |
| Drug and gene relationship at DGIdb | |||
CLDN10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction |
Expression of CLDN10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLDN10 | 9071 | 205328_at | -0.5064 | 0.8450 | |
| GSE20347 | CLDN10 | 9071 | 205328_at | -1.0465 | 0.0712 | |
| GSE23400 | CLDN10 | 9071 | 205328_at | -1.1284 | 0.0000 | |
| GSE26886 | CLDN10 | 9071 | 205328_at | -0.1509 | 0.8907 | |
| GSE29001 | CLDN10 | 9071 | 205328_at | -2.1328 | 0.0018 | |
| GSE38129 | CLDN10 | 9071 | 205328_at | -0.7869 | 0.1300 | |
| GSE45670 | CLDN10 | 9071 | 205328_at | -2.0510 | 0.0037 | |
| GSE53622 | CLDN10 | 9071 | 12728 | -0.8619 | 0.0038 | |
| GSE53624 | CLDN10 | 9071 | 12728 | -1.8996 | 0.0000 | |
| GSE63941 | CLDN10 | 9071 | 205328_at | 1.1455 | 0.4028 | |
| GSE77861 | CLDN10 | 9071 | 205328_at | -0.7654 | 0.3155 | |
| GSE97050 | CLDN10 | 9071 | A_33_P3410806 | -0.0338 | 0.9091 | |
| SRP007169 | CLDN10 | 9071 | RNAseq | -8.5575 | 0.0000 | |
| SRP064894 | CLDN10 | 9071 | RNAseq | -4.2509 | 0.0000 | |
| SRP133303 | CLDN10 | 9071 | RNAseq | -1.2917 | 0.0037 | |
| SRP159526 | CLDN10 | 9071 | RNAseq | 0.2931 | 0.7598 | |
| SRP193095 | CLDN10 | 9071 | RNAseq | -1.2049 | 0.0003 | |
| TCGA | CLDN10 | 9071 | RNAseq | 0.6019 | 0.3941 |
Upregulated datasets: 0; Downregulated datasets: 8.
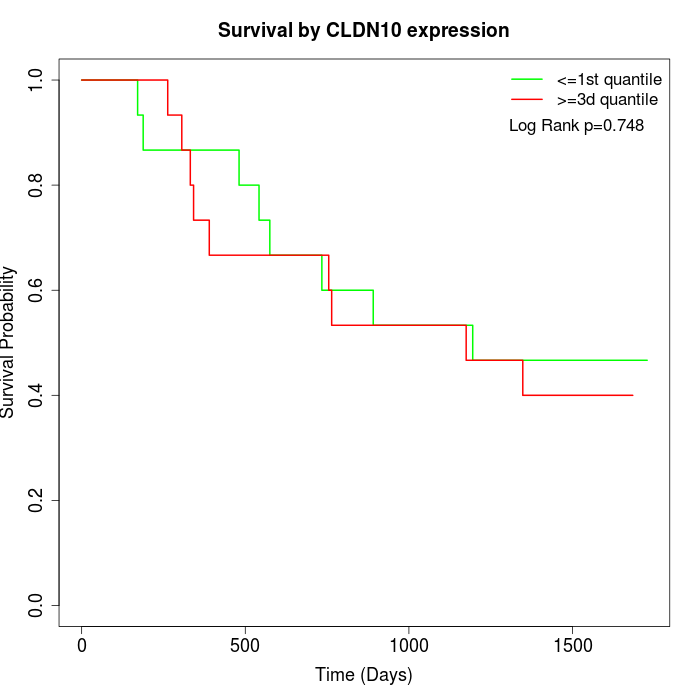
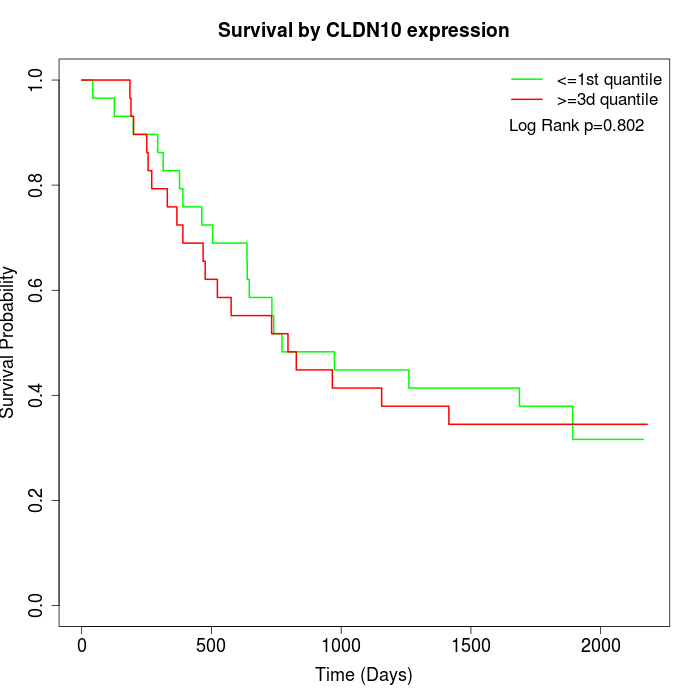
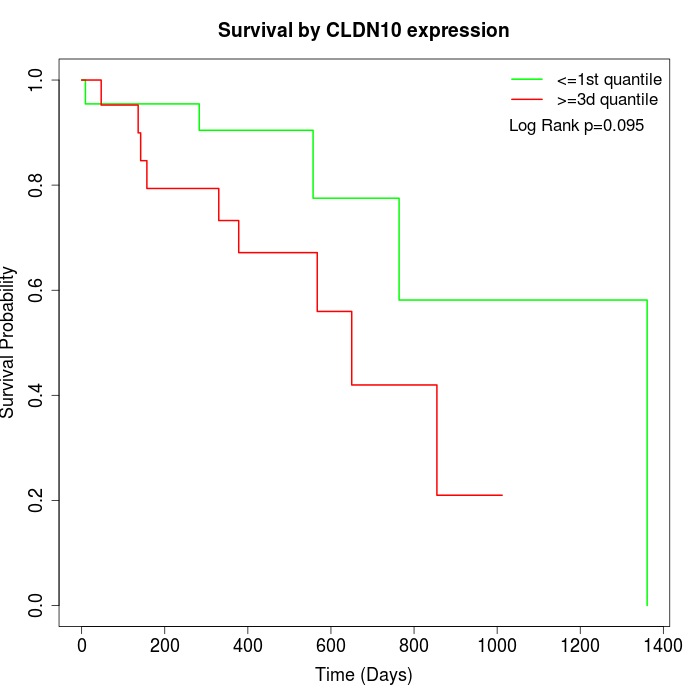
Survival by CLDN10 expression:
Note: Click image to view full size file.
Copy number change of CLDN10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLDN10 | 9071 | 4 | 9 | 17 | |
| GSE20123 | CLDN10 | 9071 | 4 | 9 | 17 | |
| GSE43470 | CLDN10 | 9071 | 2 | 12 | 29 | |
| GSE46452 | CLDN10 | 9071 | 0 | 32 | 27 | |
| GSE47630 | CLDN10 | 9071 | 3 | 27 | 10 | |
| GSE54993 | CLDN10 | 9071 | 11 | 3 | 56 | |
| GSE54994 | CLDN10 | 9071 | 3 | 13 | 37 | |
| GSE60625 | CLDN10 | 9071 | 0 | 3 | 8 | |
| GSE74703 | CLDN10 | 9071 | 2 | 10 | 24 | |
| GSE74704 | CLDN10 | 9071 | 2 | 8 | 10 | |
| TCGA | CLDN10 | 9071 | 13 | 34 | 49 |
Total number of gains: 44; Total number of losses: 160; Total Number of normals: 284.
Somatic mutations of CLDN10:
Generating mutation plots.
Highly correlated genes for CLDN10:
Showing top 20/378 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLDN10 | PRPF38A | 0.73417 | 3 | 0 | 3 |
| CLDN10 | ECD | 0.701471 | 3 | 0 | 3 |
| CLDN10 | FRMD6 | 0.691082 | 3 | 0 | 3 |
| CLDN10 | ARL16 | 0.689925 | 3 | 0 | 3 |
| CLDN10 | TMEM219 | 0.680196 | 3 | 0 | 3 |
| CLDN10 | GOLGA4 | 0.678095 | 3 | 0 | 3 |
| CLDN10 | TMEM139 | 0.674719 | 3 | 0 | 3 |
| CLDN10 | PRDX4 | 0.672986 | 3 | 0 | 3 |
| CLDN10 | MDFIC | 0.651782 | 3 | 0 | 3 |
| CLDN10 | GALNT7 | 0.643999 | 6 | 0 | 5 |
| CLDN10 | ENDOD1 | 0.643725 | 3 | 0 | 3 |
| CLDN10 | RPS9 | 0.640813 | 4 | 0 | 4 |
| CLDN10 | HBP1 | 0.639853 | 7 | 0 | 6 |
| CLDN10 | C1orf50 | 0.637866 | 3 | 0 | 3 |
| CLDN10 | PRKAR2A | 0.637265 | 4 | 0 | 4 |
| CLDN10 | ACAA1 | 0.633918 | 4 | 0 | 3 |
| CLDN10 | AKR1A1 | 0.633649 | 6 | 0 | 5 |
| CLDN10 | DHRS9 | 0.626399 | 6 | 0 | 4 |
| CLDN10 | MRPL37 | 0.626095 | 4 | 0 | 3 |
| CLDN10 | BACE2 | 0.6259 | 5 | 0 | 5 |
For details and further investigation, click here