| Full name: calcitonin receptor | Alias Symbol: CTR | ||
| Type: protein-coding gene | Cytoband: 7q21.3 | ||
| Entrez ID: 799 | HGNC ID: HGNC:1440 | Ensembl Gene: ENSG00000004948 | OMIM ID: 114131 |
| Drug and gene relationship at DGIdb | |||
Expression of CALCR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CALCR | 799 | 207887_s_at | -0.0513 | 0.8690 | |
| GSE20347 | CALCR | 799 | 207887_s_at | 0.0079 | 0.9440 | |
| GSE23400 | CALCR | 799 | 207887_s_at | -0.1288 | 0.0013 | |
| GSE26886 | CALCR | 799 | 207887_s_at | -0.2517 | 0.0039 | |
| GSE29001 | CALCR | 799 | 207887_s_at | -0.0029 | 0.9878 | |
| GSE38129 | CALCR | 799 | 207887_s_at | -0.0750 | 0.3357 | |
| GSE45670 | CALCR | 799 | 207887_s_at | 0.0138 | 0.9049 | |
| GSE53622 | CALCR | 799 | 51072 | 0.0309 | 0.7381 | |
| GSE53624 | CALCR | 799 | 51072 | -0.2180 | 0.1434 | |
| GSE63941 | CALCR | 799 | 207887_s_at | 0.0035 | 0.9859 | |
| GSE77861 | CALCR | 799 | 207887_s_at | -0.0568 | 0.7409 | |
| GSE97050 | CALCR | 799 | A_33_P3710082 | 0.0350 | 0.8926 | |
| TCGA | CALCR | 799 | RNAseq | 1.3655 | 0.0202 |
Upregulated datasets: 1; Downregulated datasets: 0.
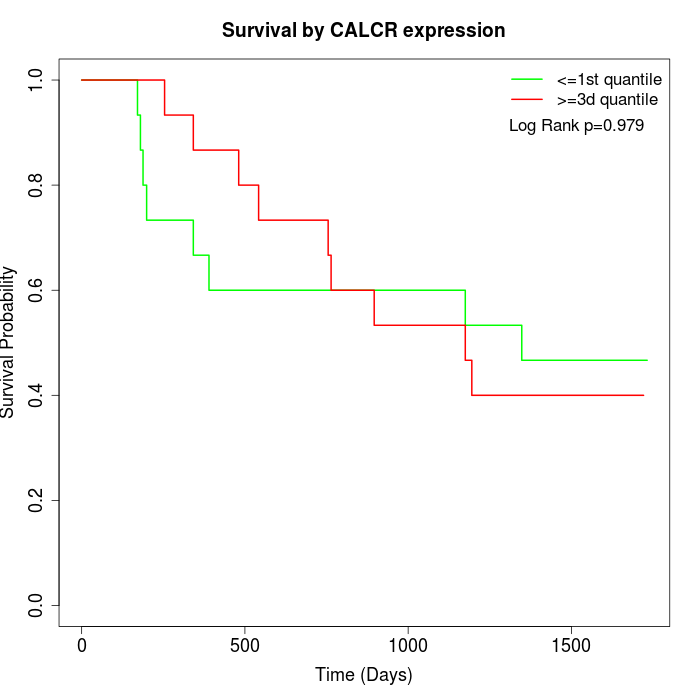
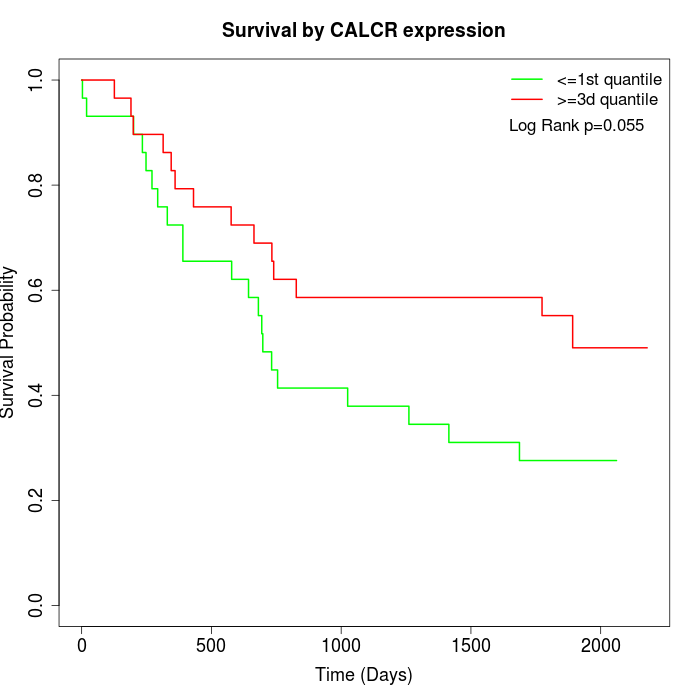
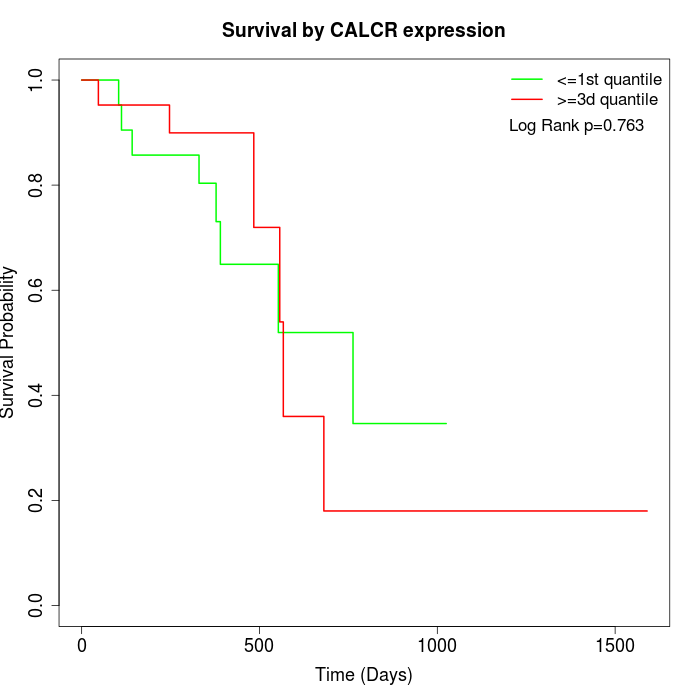
Survival by CALCR expression:
Note: Click image to view full size file.
Copy number change of CALCR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CALCR | 799 | 13 | 0 | 17 | |
| GSE20123 | CALCR | 799 | 13 | 0 | 17 | |
| GSE43470 | CALCR | 799 | 7 | 1 | 35 | |
| GSE46452 | CALCR | 799 | 12 | 0 | 47 | |
| GSE47630 | CALCR | 799 | 8 | 2 | 30 | |
| GSE54993 | CALCR | 799 | 1 | 8 | 61 | |
| GSE54994 | CALCR | 799 | 16 | 2 | 35 | |
| GSE60625 | CALCR | 799 | 0 | 0 | 11 | |
| GSE74703 | CALCR | 799 | 6 | 1 | 29 | |
| GSE74704 | CALCR | 799 | 9 | 0 | 11 | |
| TCGA | CALCR | 799 | 56 | 4 | 36 |
Total number of gains: 141; Total number of losses: 18; Total Number of normals: 329.
Somatic mutations of CALCR:
Generating mutation plots.
Highly correlated genes for CALCR:
Showing top 20/748 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CALCR | SCGN | 0.771039 | 4 | 0 | 4 |
| CALCR | CYP2D6 | 0.768849 | 5 | 0 | 5 |
| CALCR | KIR3DL3 | 0.754366 | 5 | 0 | 5 |
| CALCR | PHOSPHO1 | 0.742853 | 3 | 0 | 3 |
| CALCR | HMOX1 | 0.737632 | 3 | 0 | 3 |
| CALCR | GPR39 | 0.735228 | 3 | 0 | 3 |
| CALCR | TERT | 0.73188 | 5 | 0 | 5 |
| CALCR | GYPB | 0.726137 | 5 | 0 | 5 |
| CALCR | CDH2 | 0.725574 | 3 | 0 | 3 |
| CALCR | PRAMEF12 | 0.723147 | 7 | 0 | 7 |
| CALCR | SLC10A1 | 0.721171 | 6 | 0 | 5 |
| CALCR | SLC5A4 | 0.716713 | 6 | 0 | 5 |
| CALCR | CLCNKB | 0.716272 | 3 | 0 | 3 |
| CALCR | AICDA | 0.714686 | 4 | 0 | 4 |
| CALCR | CEMP1 | 0.713428 | 4 | 0 | 3 |
| CALCR | GAD2 | 0.711707 | 5 | 0 | 5 |
| CALCR | KRT77 | 0.710044 | 3 | 0 | 3 |
| CALCR | FTHL17 | 0.706577 | 3 | 0 | 3 |
| CALCR | ABCC6 | 0.704553 | 4 | 0 | 3 |
| CALCR | NCAN | 0.70386 | 6 | 0 | 5 |
For details and further investigation, click here