| Full name: growth differentiation factor 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 9573 | HGNC ID: HGNC:4218 | Ensembl Gene: ENSG00000184344 | OMIM ID: 606522 |
| Drug and gene relationship at DGIdb | |||
Expression of GDF3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GDF3 | 9573 | 220053_at | -0.0383 | 0.8643 | |
| GSE20347 | GDF3 | 9573 | 220053_at | -0.0428 | 0.5392 | |
| GSE23400 | GDF3 | 9573 | 220053_at | -0.1809 | 0.0000 | |
| GSE26886 | GDF3 | 9573 | 220053_at | -0.2040 | 0.1327 | |
| GSE29001 | GDF3 | 9573 | 220053_at | -0.2714 | 0.0773 | |
| GSE38129 | GDF3 | 9573 | 220053_at | -0.1166 | 0.0600 | |
| GSE45670 | GDF3 | 9573 | 220053_at | 0.0731 | 0.3557 | |
| GSE53622 | GDF3 | 9573 | 107196 | 0.0880 | 0.5062 | |
| GSE53624 | GDF3 | 9573 | 107196 | 0.0223 | 0.8305 | |
| GSE63941 | GDF3 | 9573 | 220053_at | 0.1922 | 0.1734 | |
| GSE77861 | GDF3 | 9573 | 220053_at | -0.0737 | 0.5482 | |
| TCGA | GDF3 | 9573 | RNAseq | 1.9420 | 0.0417 |
Upregulated datasets: 1; Downregulated datasets: 0.
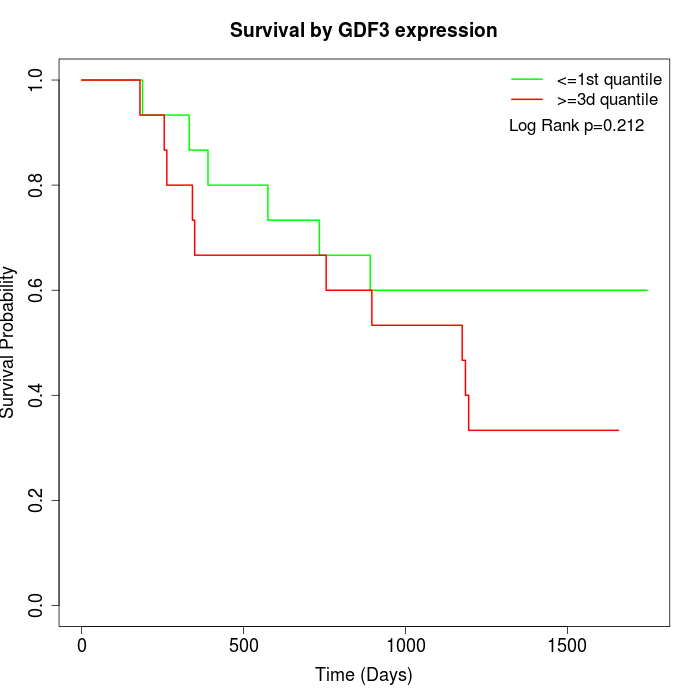
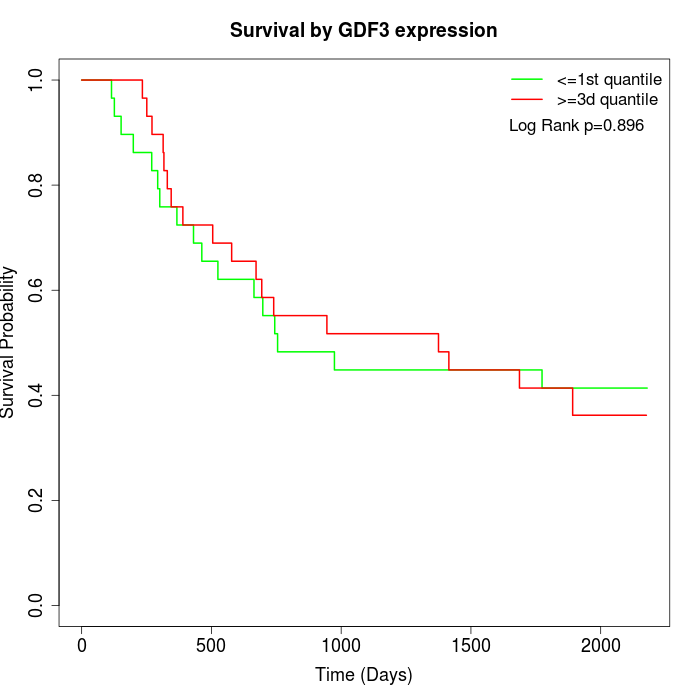
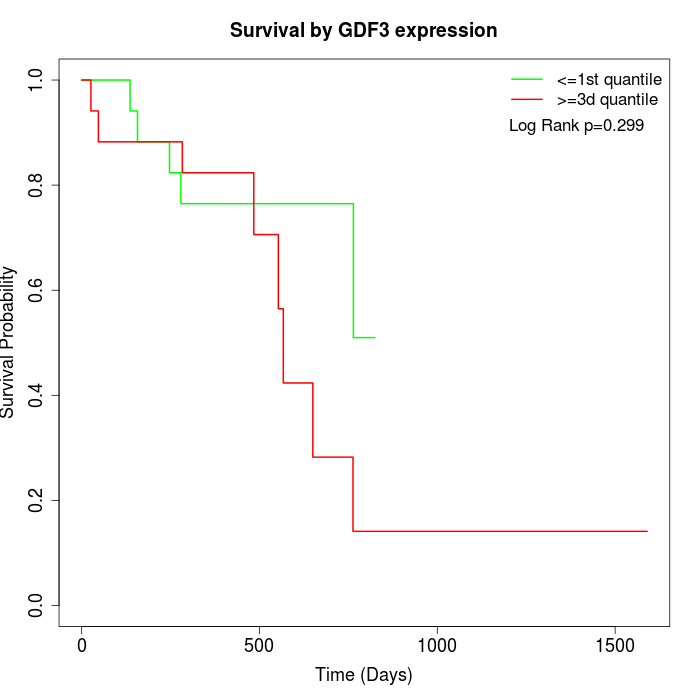
Survival by GDF3 expression:
Note: Click image to view full size file.
Copy number change of GDF3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GDF3 | 9573 | 6 | 3 | 21 | |
| GSE20123 | GDF3 | 9573 | 6 | 3 | 21 | |
| GSE43470 | GDF3 | 9573 | 7 | 3 | 33 | |
| GSE46452 | GDF3 | 9573 | 10 | 1 | 48 | |
| GSE47630 | GDF3 | 9573 | 12 | 2 | 26 | |
| GSE54993 | GDF3 | 9573 | 1 | 10 | 59 | |
| GSE54994 | GDF3 | 9573 | 9 | 5 | 39 | |
| GSE60625 | GDF3 | 9573 | 0 | 1 | 10 | |
| GSE74703 | GDF3 | 9573 | 7 | 2 | 27 | |
| GSE74704 | GDF3 | 9573 | 4 | 2 | 14 | |
| TCGA | GDF3 | 9573 | 40 | 6 | 50 |
Total number of gains: 102; Total number of losses: 38; Total Number of normals: 348.
Somatic mutations of GDF3:
Generating mutation plots.
Highly correlated genes for GDF3:
Showing top 20/875 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GDF3 | SLC4A4 | 0.750518 | 4 | 0 | 4 |
| GDF3 | RHAG | 0.739731 | 3 | 0 | 3 |
| GDF3 | CPN1 | 0.721127 | 4 | 0 | 4 |
| GDF3 | CNR2 | 0.709318 | 5 | 0 | 5 |
| GDF3 | TBX5 | 0.706248 | 4 | 0 | 4 |
| GDF3 | CGA | 0.706011 | 4 | 0 | 4 |
| GDF3 | KCNJ10 | 0.688682 | 6 | 0 | 6 |
| GDF3 | CTRC | 0.688647 | 3 | 0 | 3 |
| GDF3 | ADM2 | 0.687468 | 5 | 0 | 5 |
| GDF3 | CBL | 0.684842 | 4 | 0 | 4 |
| GDF3 | HAND1 | 0.683779 | 5 | 0 | 5 |
| GDF3 | PRB1 | 0.679829 | 6 | 0 | 6 |
| GDF3 | EDA2R | 0.672511 | 5 | 0 | 4 |
| GDF3 | NOX5 | 0.67184 | 5 | 0 | 4 |
| GDF3 | RSPH6A | 0.671431 | 6 | 0 | 6 |
| GDF3 | ART1 | 0.671428 | 6 | 0 | 5 |
| GDF3 | PTPRO | 0.670939 | 3 | 0 | 3 |
| GDF3 | NCR1 | 0.670124 | 5 | 0 | 4 |
| GDF3 | ADRB3 | 0.668286 | 3 | 0 | 3 |
| GDF3 | TCTN2 | 0.667381 | 3 | 0 | 3 |
For details and further investigation, click here