| Full name: complexin 2 | Alias Symbol: CPX-2|DKFZp547D155 | ||
| Type: protein-coding gene | Cytoband: 5q35.2 | ||
| Entrez ID: 10814 | HGNC ID: HGNC:2310 | Ensembl Gene: ENSG00000145920 | OMIM ID: 605033 |
| Drug and gene relationship at DGIdb | |||
Expression of CPLX2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CPLX2 | 10814 | 225815_at | -0.1008 | 0.6759 | |
| GSE20347 | CPLX2 | 10814 | 206368_at | -0.0439 | 0.5054 | |
| GSE23400 | CPLX2 | 10814 | 206368_at | -0.0471 | 0.0262 | |
| GSE26886 | CPLX2 | 10814 | 225815_at | 0.1764 | 0.1127 | |
| GSE29001 | CPLX2 | 10814 | 206368_at | -0.1169 | 0.3144 | |
| GSE38129 | CPLX2 | 10814 | 206368_at | -0.0575 | 0.2779 | |
| GSE45670 | CPLX2 | 10814 | 206368_at | -0.0228 | 0.8512 | |
| GSE53622 | CPLX2 | 10814 | 53798 | 0.1042 | 0.2717 | |
| GSE53624 | CPLX2 | 10814 | 53798 | 0.0820 | 0.4346 | |
| GSE63941 | CPLX2 | 10814 | 225815_at | 0.0529 | 0.7270 | |
| GSE77861 | CPLX2 | 10814 | 225815_at | -0.0009 | 0.9947 | |
| GSE97050 | CPLX2 | 10814 | A_24_P391868 | 0.0583 | 0.8410 | |
| TCGA | CPLX2 | 10814 | RNAseq | -1.5487 | 0.0626 |
Upregulated datasets: 0; Downregulated datasets: 0.
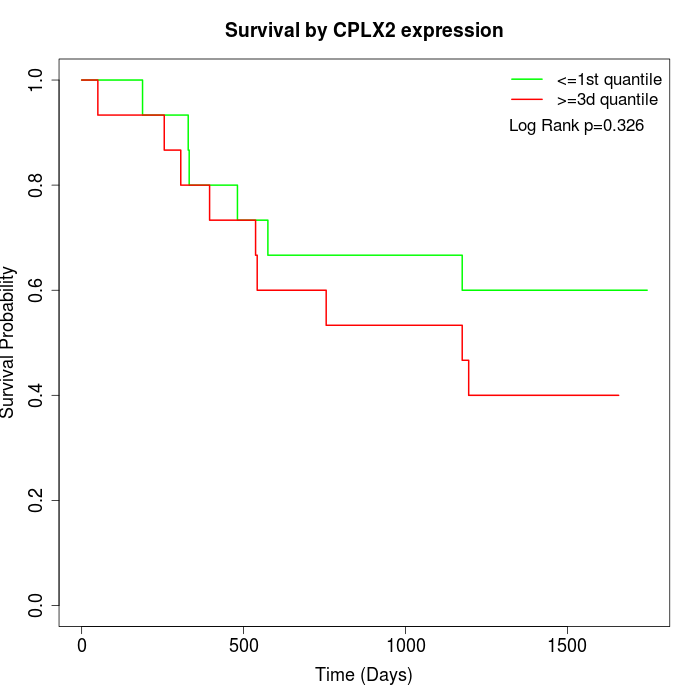
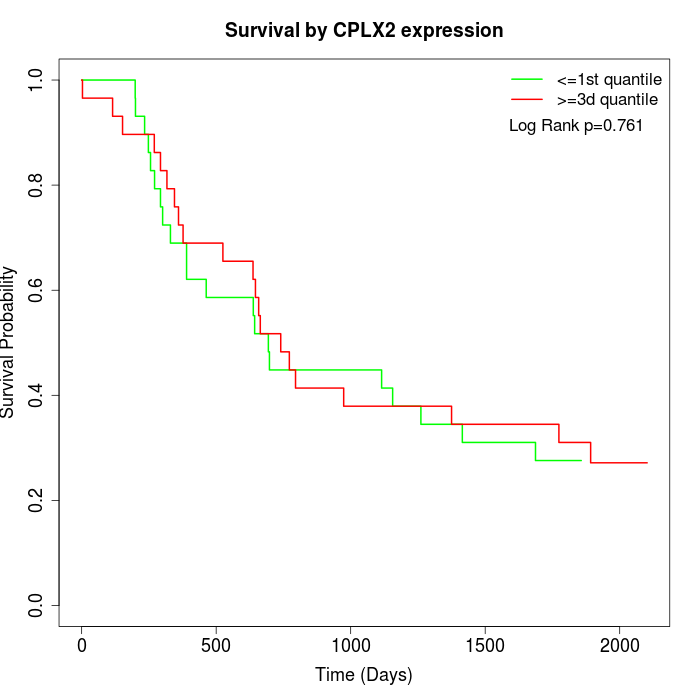
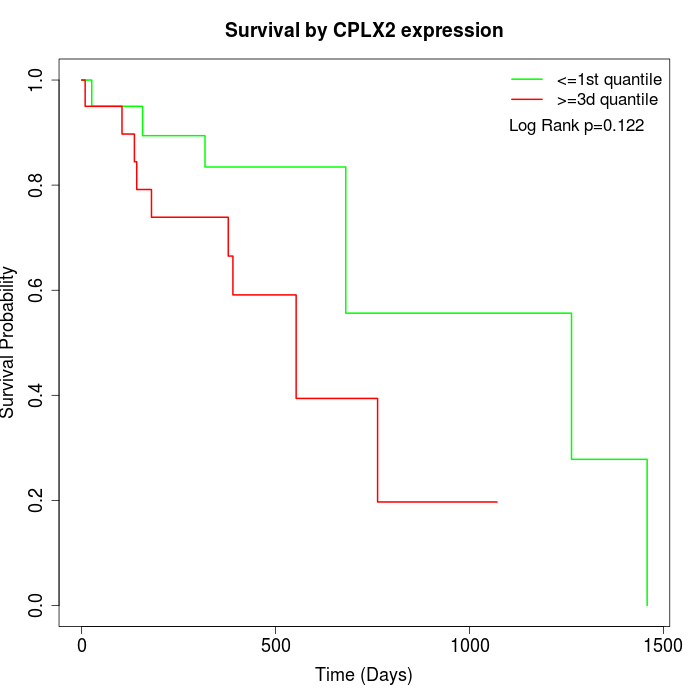
Survival by CPLX2 expression:
Note: Click image to view full size file.
Copy number change of CPLX2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CPLX2 | 10814 | 3 | 12 | 15 | |
| GSE20123 | CPLX2 | 10814 | 3 | 12 | 15 | |
| GSE43470 | CPLX2 | 10814 | 1 | 10 | 32 | |
| GSE46452 | CPLX2 | 10814 | 0 | 27 | 32 | |
| GSE47630 | CPLX2 | 10814 | 0 | 20 | 20 | |
| GSE54993 | CPLX2 | 10814 | 10 | 2 | 58 | |
| GSE54994 | CPLX2 | 10814 | 2 | 17 | 34 | |
| GSE60625 | CPLX2 | 10814 | 1 | 0 | 10 | |
| GSE74703 | CPLX2 | 10814 | 1 | 7 | 28 | |
| GSE74704 | CPLX2 | 10814 | 2 | 5 | 13 | |
| TCGA | CPLX2 | 10814 | 7 | 35 | 54 |
Total number of gains: 30; Total number of losses: 147; Total Number of normals: 311.
Somatic mutations of CPLX2:
Generating mutation plots.
Highly correlated genes for CPLX2:
Showing top 20/726 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CPLX2 | YY2 | 0.881236 | 3 | 0 | 3 |
| CPLX2 | PAX6 | 0.879005 | 3 | 0 | 3 |
| CPLX2 | DCDC1 | 0.872747 | 3 | 0 | 3 |
| CPLX2 | ZNF763 | 0.852351 | 3 | 0 | 3 |
| CPLX2 | PRSS38 | 0.847634 | 3 | 0 | 3 |
| CPLX2 | OR2T27 | 0.845407 | 3 | 0 | 3 |
| CPLX2 | C1QL2 | 0.830098 | 3 | 0 | 3 |
| CPLX2 | OR2T1 | 0.817177 | 3 | 0 | 3 |
| CPLX2 | BBC3 | 0.81489 | 3 | 0 | 3 |
| CPLX2 | C9orf153 | 0.811114 | 3 | 0 | 3 |
| CPLX2 | PLA2G2C | 0.810764 | 3 | 0 | 3 |
| CPLX2 | STRC | 0.810132 | 3 | 0 | 3 |
| CPLX2 | MRGPRG | 0.804553 | 3 | 0 | 3 |
| CPLX2 | KRTAP10-12 | 0.795096 | 3 | 0 | 3 |
| CPLX2 | ZAN | 0.794305 | 3 | 0 | 3 |
| CPLX2 | OR10AD1 | 0.793707 | 3 | 0 | 3 |
| CPLX2 | KRTAP2-1 | 0.793588 | 3 | 0 | 3 |
| CPLX2 | PDZRN3 | 0.789954 | 3 | 0 | 3 |
| CPLX2 | FAM71F1 | 0.788992 | 3 | 0 | 3 |
| CPLX2 | OTOG | 0.786956 | 3 | 0 | 3 |
For details and further investigation, click here