| Full name: C-terminal binding protein 2 | Alias Symbol: ribeye | ||
| Type: protein-coding gene | Cytoband: 10q26.13 | ||
| Entrez ID: 1488 | HGNC ID: HGNC:2495 | Ensembl Gene: ENSG00000175029 | OMIM ID: 602619 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CTBP2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04310 | Wnt signaling pathway | |
| hsa04330 | Notch signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05220 | Chronic myeloid leukemia |
Expression of CTBP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTBP2 | 1488 | 201220_x_at | 0.4519 | 0.3586 | |
| GSE20347 | CTBP2 | 1488 | 210835_s_at | 0.6877 | 0.0004 | |
| GSE23400 | CTBP2 | 1488 | 201220_x_at | 0.1725 | 0.0039 | |
| GSE26886 | CTBP2 | 1488 | 201220_x_at | 0.8814 | 0.0000 | |
| GSE29001 | CTBP2 | 1488 | 210835_s_at | 0.3550 | 0.0780 | |
| GSE38129 | CTBP2 | 1488 | 201220_x_at | 0.3486 | 0.0436 | |
| GSE45670 | CTBP2 | 1488 | 201220_x_at | 0.1666 | 0.1167 | |
| GSE53622 | CTBP2 | 1488 | 84972 | 0.2567 | 0.0000 | |
| GSE53624 | CTBP2 | 1488 | 80584 | 0.4267 | 0.0000 | |
| GSE63941 | CTBP2 | 1488 | 201218_at | 0.4885 | 0.0943 | |
| GSE77861 | CTBP2 | 1488 | 201220_x_at | 0.4417 | 0.0547 | |
| GSE97050 | CTBP2 | 1488 | A_24_P103922 | 0.2881 | 0.2465 | |
| SRP007169 | CTBP2 | 1488 | RNAseq | 0.8204 | 0.0750 | |
| SRP008496 | CTBP2 | 1488 | RNAseq | 1.0329 | 0.0021 | |
| SRP064894 | CTBP2 | 1488 | RNAseq | -0.1911 | 0.0223 | |
| SRP133303 | CTBP2 | 1488 | RNAseq | 0.1136 | 0.3735 | |
| SRP159526 | CTBP2 | 1488 | RNAseq | -0.0784 | 0.7392 | |
| SRP193095 | CTBP2 | 1488 | RNAseq | 0.2144 | 0.0385 | |
| SRP219564 | CTBP2 | 1488 | RNAseq | -0.1127 | 0.6838 | |
| TCGA | CTBP2 | 1488 | RNAseq | -0.1246 | 0.0051 |
Upregulated datasets: 1; Downregulated datasets: 0.
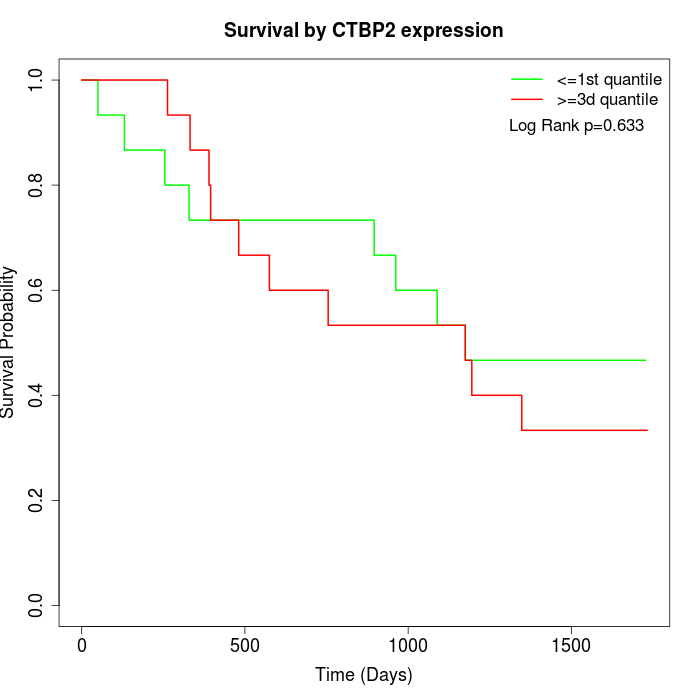
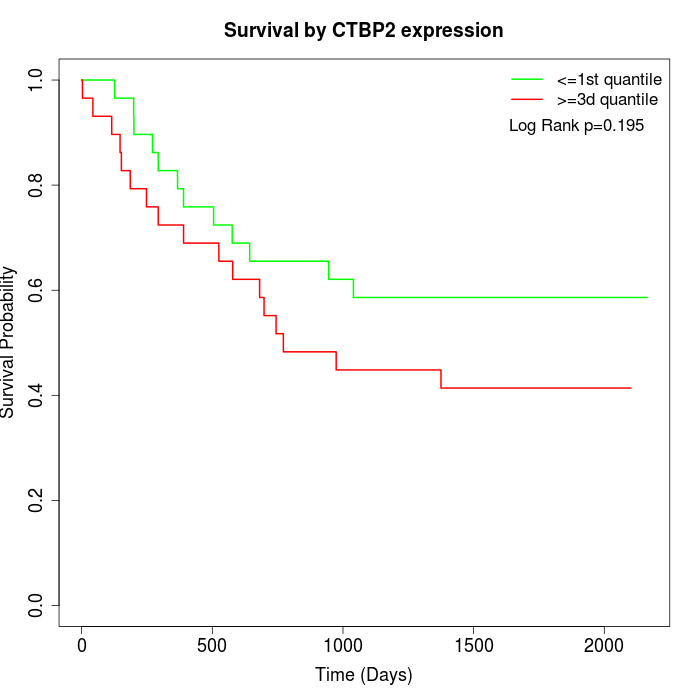
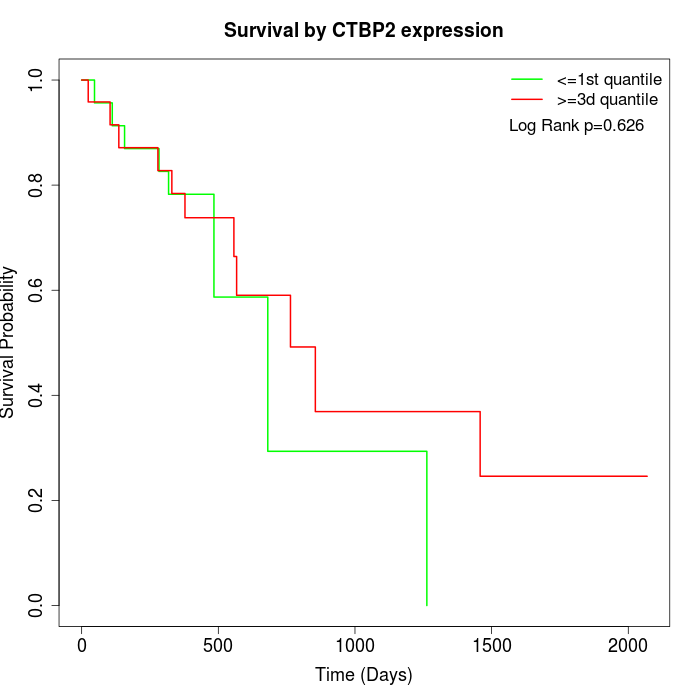
Survival by CTBP2 expression:
Note: Click image to view full size file.
Copy number change of CTBP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTBP2 | 1488 | 2 | 7 | 21 | |
| GSE20123 | CTBP2 | 1488 | 1 | 7 | 22 | |
| GSE43470 | CTBP2 | 1488 | 0 | 10 | 33 | |
| GSE46452 | CTBP2 | 1488 | 0 | 11 | 48 | |
| GSE47630 | CTBP2 | 1488 | 2 | 14 | 24 | |
| GSE54993 | CTBP2 | 1488 | 8 | 1 | 61 | |
| GSE54994 | CTBP2 | 1488 | 1 | 9 | 43 | |
| GSE60625 | CTBP2 | 1488 | 0 | 0 | 11 | |
| GSE74703 | CTBP2 | 1488 | 0 | 6 | 30 | |
| GSE74704 | CTBP2 | 1488 | 1 | 2 | 17 | |
| TCGA | CTBP2 | 1488 | 6 | 26 | 64 |
Total number of gains: 21; Total number of losses: 93; Total Number of normals: 374.
Somatic mutations of CTBP2:
Generating mutation plots.
Highly correlated genes for CTBP2:
Showing top 20/1213 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTBP2 | TCTN3 | 0.823782 | 3 | 0 | 3 |
| CTBP2 | RNASEH2C | 0.801655 | 3 | 0 | 3 |
| CTBP2 | HYLS1 | 0.798247 | 3 | 0 | 3 |
| CTBP2 | NOM1 | 0.794186 | 4 | 0 | 4 |
| CTBP2 | UBB | 0.776943 | 3 | 0 | 3 |
| CTBP2 | PRR12 | 0.770548 | 3 | 0 | 3 |
| CTBP2 | WDR81 | 0.770093 | 3 | 0 | 3 |
| CTBP2 | MRPL9 | 0.769786 | 3 | 0 | 3 |
| CTBP2 | DHRSX | 0.768588 | 3 | 0 | 3 |
| CTBP2 | LRRC34 | 0.766721 | 3 | 0 | 3 |
| CTBP2 | NUFIP1 | 0.765992 | 3 | 0 | 3 |
| CTBP2 | ZNF567 | 0.763712 | 3 | 0 | 3 |
| CTBP2 | TRMT6 | 0.760795 | 3 | 0 | 3 |
| CTBP2 | SLC39A3 | 0.760396 | 3 | 0 | 3 |
| CTBP2 | METAP1D | 0.760361 | 3 | 0 | 3 |
| CTBP2 | NAF1 | 0.757201 | 3 | 0 | 3 |
| CTBP2 | MRPS9 | 0.756266 | 3 | 0 | 3 |
| CTBP2 | SLC30A7 | 0.751444 | 4 | 0 | 3 |
| CTBP2 | POMZP3 | 0.749226 | 4 | 0 | 4 |
| CTBP2 | INPP4A | 0.748856 | 3 | 0 | 3 |
For details and further investigation, click here