| Full name: cytosolic thiouridylase subunit 1 | Alias Symbol: MGC17332|NCS6 | ||
| Type: protein-coding gene | Cytoband: 19q13.41 | ||
| Entrez ID: 90353 | HGNC ID: HGNC:29590 | Ensembl Gene: ENSG00000142544 | OMIM ID: 612694 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CTU1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | CTU1 | 90353 | 72860 | -0.1756 | 0.0000 | |
| GSE53624 | CTU1 | 90353 | 72860 | -0.0478 | 0.3072 | |
| GSE97050 | CTU1 | 90353 | A_33_P3395976 | -0.0064 | 0.9795 | |
| SRP007169 | CTU1 | 90353 | RNAseq | -0.3777 | 0.3981 | |
| SRP008496 | CTU1 | 90353 | RNAseq | -0.7121 | 0.0418 | |
| SRP064894 | CTU1 | 90353 | RNAseq | 0.9852 | 0.0266 | |
| SRP133303 | CTU1 | 90353 | RNAseq | 0.2294 | 0.2514 | |
| SRP159526 | CTU1 | 90353 | RNAseq | 0.5690 | 0.1501 | |
| SRP193095 | CTU1 | 90353 | RNAseq | 0.4761 | 0.0008 | |
| SRP219564 | CTU1 | 90353 | RNAseq | 0.2521 | 0.4921 | |
| TCGA | CTU1 | 90353 | RNAseq | 0.2083 | 0.0046 |
Upregulated datasets: 0; Downregulated datasets: 0.
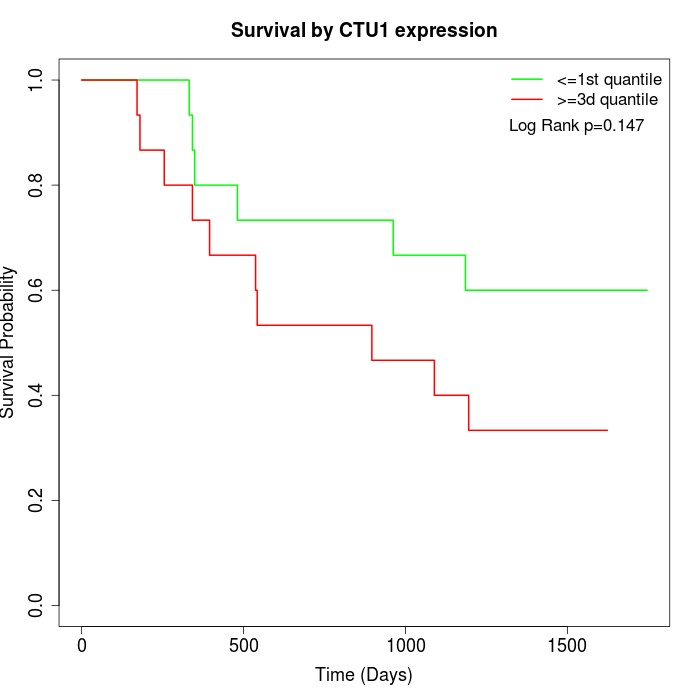
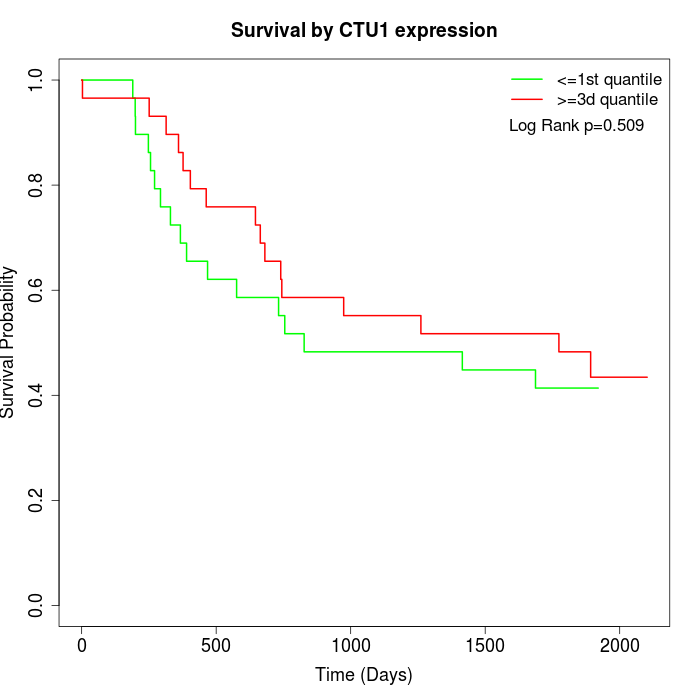
Survival by CTU1 expression:
Note: Click image to view full size file.
Copy number change of CTU1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTU1 | 90353 | 3 | 4 | 23 | |
| GSE20123 | CTU1 | 90353 | 3 | 3 | 24 | |
| GSE43470 | CTU1 | 90353 | 4 | 11 | 28 | |
| GSE46452 | CTU1 | 90353 | 45 | 1 | 13 | |
| GSE47630 | CTU1 | 90353 | 9 | 6 | 25 | |
| GSE54993 | CTU1 | 90353 | 17 | 4 | 49 | |
| GSE54994 | CTU1 | 90353 | 4 | 14 | 35 | |
| GSE60625 | CTU1 | 90353 | 9 | 0 | 2 | |
| GSE74703 | CTU1 | 90353 | 4 | 7 | 25 | |
| GSE74704 | CTU1 | 90353 | 3 | 1 | 16 | |
| TCGA | CTU1 | 90353 | 14 | 19 | 63 |
Total number of gains: 115; Total number of losses: 70; Total Number of normals: 303.
Somatic mutations of CTU1:
Generating mutation plots.
Highly correlated genes for CTU1:
Showing top 20/30 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTU1 | BHLHA9 | 0.750576 | 3 | 0 | 3 |
| CTU1 | SLC22A7 | 0.744588 | 3 | 0 | 3 |
| CTU1 | POLD1 | 0.743055 | 3 | 0 | 3 |
| CTU1 | TTC7A | 0.736748 | 3 | 0 | 3 |
| CTU1 | C8G | 0.732351 | 3 | 0 | 3 |
| CTU1 | SAMD4A | 0.728002 | 3 | 0 | 3 |
| CTU1 | ZBTB45 | 0.725107 | 3 | 0 | 3 |
| CTU1 | MUC2 | 0.724105 | 3 | 0 | 3 |
| CTU1 | SLC19A3 | 0.722156 | 3 | 0 | 3 |
| CTU1 | ANKRD33 | 0.720462 | 3 | 0 | 3 |
| CTU1 | GDF7 | 0.714799 | 3 | 0 | 3 |
| CTU1 | GPR150 | 0.711157 | 3 | 0 | 3 |
| CTU1 | ZNF329 | 0.70722 | 3 | 0 | 3 |
| CTU1 | NPAS1 | 0.706863 | 3 | 0 | 3 |
| CTU1 | ANKRD33B | 0.704939 | 3 | 0 | 3 |
| CTU1 | ESPNL | 0.702941 | 3 | 0 | 3 |
| CTU1 | MAMSTR | 0.699351 | 3 | 0 | 3 |
| CTU1 | TMUB1 | 0.698374 | 3 | 0 | 3 |
| CTU1 | GATA2 | 0.6969 | 3 | 0 | 3 |
| CTU1 | RETN | 0.695668 | 3 | 0 | 3 |
For details and further investigation, click here