| Full name: G protein-coupled receptor 150 | Alias Symbol: PGR11 | ||
| Type: protein-coding gene | Cytoband: 5q15 | ||
| Entrez ID: 285601 | HGNC ID: HGNC:23628 | Ensembl Gene: ENSG00000178015 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GPR150:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GPR150 | 285601 | 231349_at | 0.1806 | 0.5376 | |
| GSE26886 | GPR150 | 285601 | 231349_at | 0.3413 | 0.0061 | |
| GSE45670 | GPR150 | 285601 | 231349_at | 0.0958 | 0.3443 | |
| GSE53622 | GPR150 | 285601 | 19897 | -0.1399 | 0.0323 | |
| GSE53624 | GPR150 | 285601 | 19897 | -0.0457 | 0.6057 | |
| GSE63941 | GPR150 | 285601 | 231349_at | 0.2170 | 0.2986 | |
| GSE77861 | GPR150 | 285601 | 231349_at | -0.0323 | 0.8202 | |
| GSE97050 | GPR150 | 285601 | A_24_P37887 | 0.0124 | 0.9611 | |
| TCGA | GPR150 | 285601 | RNAseq | -1.2335 | 0.0039 |
Upregulated datasets: 0; Downregulated datasets: 1.
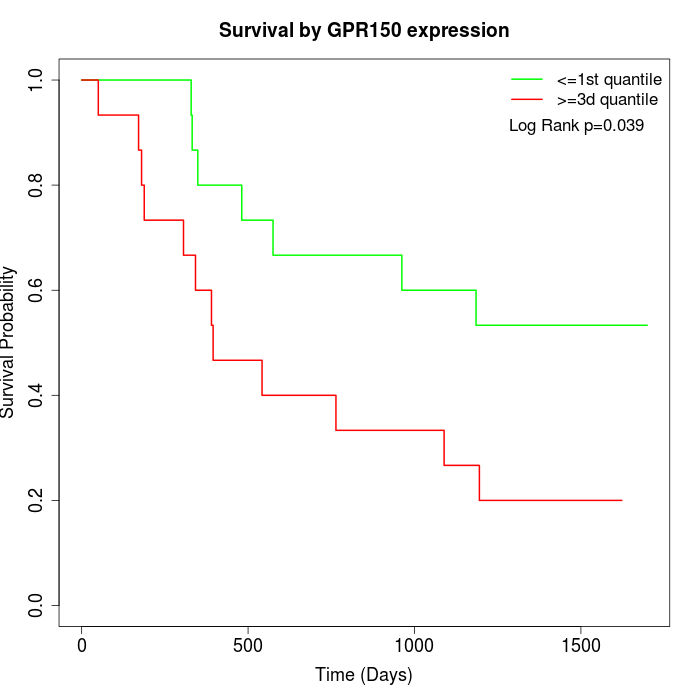
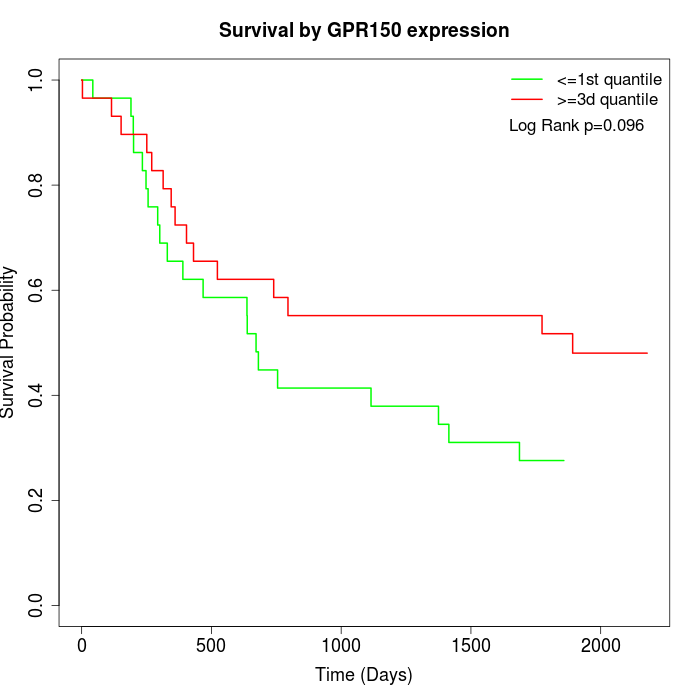
Survival by GPR150 expression:
Note: Click image to view full size file.
Copy number change of GPR150:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GPR150 | 285601 | 1 | 13 | 16 | |
| GSE20123 | GPR150 | 285601 | 1 | 13 | 16 | |
| GSE43470 | GPR150 | 285601 | 3 | 9 | 31 | |
| GSE46452 | GPR150 | 285601 | 0 | 28 | 31 | |
| GSE47630 | GPR150 | 285601 | 0 | 20 | 20 | |
| GSE54993 | GPR150 | 285601 | 8 | 1 | 61 | |
| GSE54994 | GPR150 | 285601 | 1 | 18 | 34 | |
| GSE60625 | GPR150 | 285601 | 0 | 0 | 11 | |
| GSE74703 | GPR150 | 285601 | 2 | 7 | 27 | |
| GSE74704 | GPR150 | 285601 | 1 | 7 | 12 | |
| TCGA | GPR150 | 285601 | 3 | 44 | 49 |
Total number of gains: 20; Total number of losses: 160; Total Number of normals: 308.
Somatic mutations of GPR150:
Generating mutation plots.
Highly correlated genes for GPR150:
Showing top 20/292 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GPR150 | ASPHD1 | 0.851094 | 3 | 0 | 3 |
| GPR150 | LDHC | 0.850059 | 3 | 0 | 3 |
| GPR150 | FAM177B | 0.805735 | 3 | 0 | 3 |
| GPR150 | SHISA8 | 0.795488 | 3 | 0 | 3 |
| GPR150 | C19orf81 | 0.786118 | 3 | 0 | 3 |
| GPR150 | PIP5KL1 | 0.783736 | 3 | 0 | 3 |
| GPR150 | SLC22A7 | 0.779421 | 4 | 0 | 4 |
| GPR150 | GDF7 | 0.777854 | 3 | 0 | 3 |
| GPR150 | CCNJL | 0.77628 | 3 | 0 | 3 |
| GPR150 | WNT9A | 0.766493 | 3 | 0 | 3 |
| GPR150 | MAPK15 | 0.766463 | 3 | 0 | 3 |
| GPR150 | COPS7B | 0.76286 | 3 | 0 | 3 |
| GPR150 | KCNC3 | 0.75954 | 3 | 0 | 3 |
| GPR150 | EVX1 | 0.756235 | 5 | 0 | 5 |
| GPR150 | CABP4 | 0.754578 | 3 | 0 | 3 |
| GPR150 | CORT | 0.751318 | 3 | 0 | 3 |
| GPR150 | SRRM3 | 0.750949 | 3 | 0 | 3 |
| GPR150 | SP8 | 0.750744 | 4 | 0 | 4 |
| GPR150 | ZNF497 | 0.74637 | 3 | 0 | 3 |
| GPR150 | TP73 | 0.745286 | 4 | 0 | 3 |
For details and further investigation, click here