| Full name: cullin 4B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xq24 | ||
| Entrez ID: 8450 | HGNC ID: HGNC:2555 | Ensembl Gene: ENSG00000158290 | OMIM ID: 300304 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CUL4B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CUL4B | 8450 | 202214_s_at | -0.5506 | 0.1411 | |
| GSE20347 | CUL4B | 8450 | 202214_s_at | -0.5967 | 0.0001 | |
| GSE23400 | CUL4B | 8450 | 202214_s_at | -0.2874 | 0.0000 | |
| GSE26886 | CUL4B | 8450 | 202214_s_at | -0.5239 | 0.0125 | |
| GSE29001 | CUL4B | 8450 | 202214_s_at | -0.3846 | 0.2026 | |
| GSE38129 | CUL4B | 8450 | 202214_s_at | -0.4900 | 0.0000 | |
| GSE45670 | CUL4B | 8450 | 202214_s_at | -0.2887 | 0.0718 | |
| GSE53622 | CUL4B | 8450 | 54129 | -0.4983 | 0.0000 | |
| GSE53624 | CUL4B | 8450 | 54129 | -0.8100 | 0.0000 | |
| GSE63941 | CUL4B | 8450 | 202213_s_at | 0.1286 | 0.8344 | |
| GSE77861 | CUL4B | 8450 | 202214_s_at | -0.4123 | 0.0422 | |
| GSE97050 | CUL4B | 8450 | A_23_P422178 | -0.1903 | 0.4135 | |
| SRP007169 | CUL4B | 8450 | RNAseq | -1.3965 | 0.0004 | |
| SRP008496 | CUL4B | 8450 | RNAseq | -0.7747 | 0.0260 | |
| SRP064894 | CUL4B | 8450 | RNAseq | -0.7420 | 0.0000 | |
| SRP133303 | CUL4B | 8450 | RNAseq | -0.2149 | 0.3305 | |
| SRP159526 | CUL4B | 8450 | RNAseq | -0.6421 | 0.0010 | |
| SRP193095 | CUL4B | 8450 | RNAseq | -0.9020 | 0.0000 | |
| SRP219564 | CUL4B | 8450 | RNAseq | -0.6542 | 0.0183 | |
| TCGA | CUL4B | 8450 | RNAseq | -0.0091 | 0.8371 |
Upregulated datasets: 0; Downregulated datasets: 1.
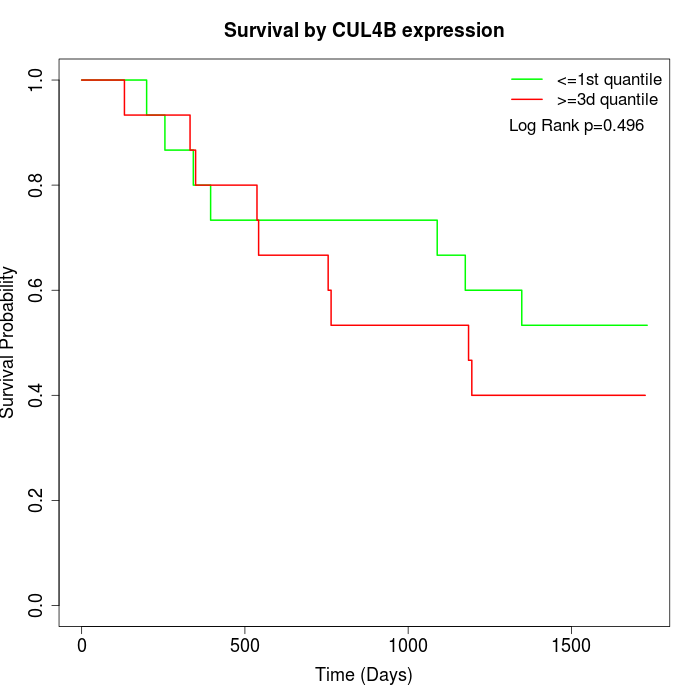
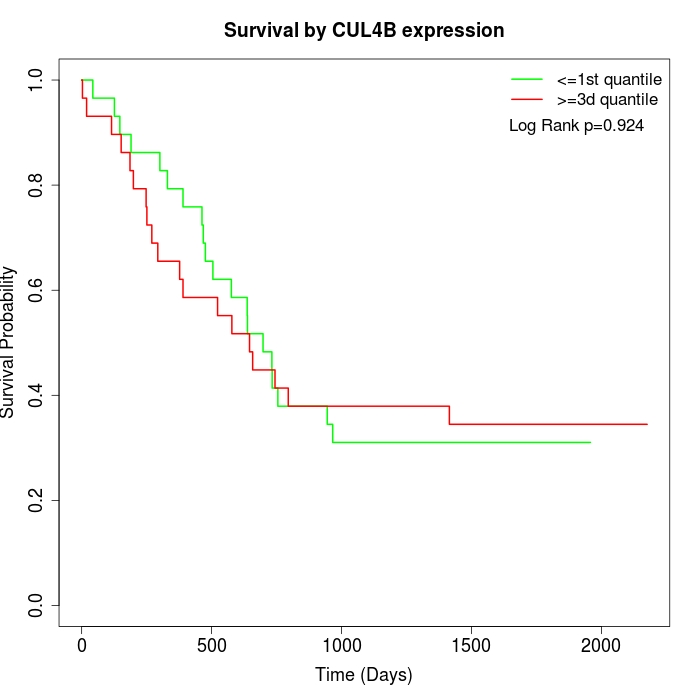
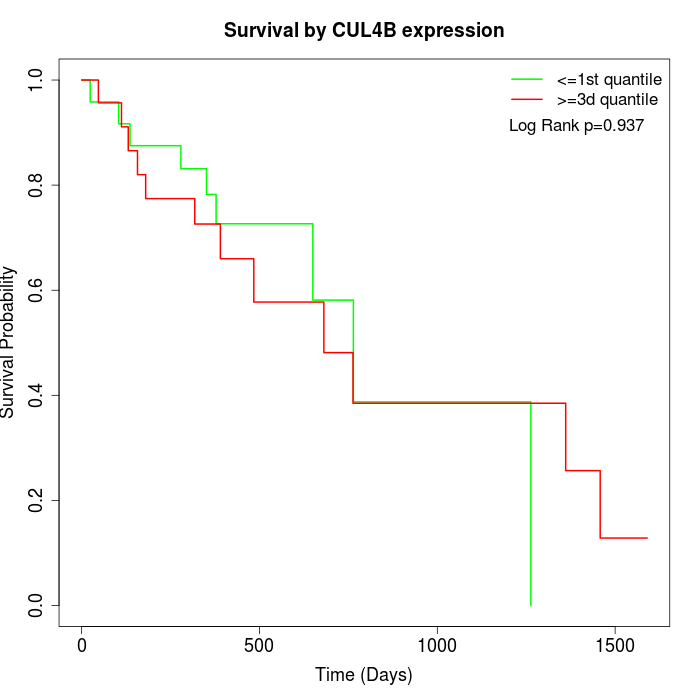
Survival by CUL4B expression:
Note: Click image to view full size file.
Copy number change of CUL4B:
No record found for this gene.
Somatic mutations of CUL4B:
Generating mutation plots.
Highly correlated genes for CUL4B:
Showing top 20/1493 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CUL4B | ZCCHC9 | 0.801005 | 3 | 0 | 3 |
| CUL4B | CYP4V2 | 0.800672 | 3 | 0 | 3 |
| CUL4B | MKLN1 | 0.792881 | 3 | 0 | 3 |
| CUL4B | TMEM161B | 0.785828 | 3 | 0 | 3 |
| CUL4B | FBXL3 | 0.780592 | 5 | 0 | 5 |
| CUL4B | ECHDC1 | 0.779432 | 3 | 0 | 3 |
| CUL4B | MIER3 | 0.776689 | 3 | 0 | 3 |
| CUL4B | ZBED5 | 0.765726 | 3 | 0 | 3 |
| CUL4B | ANKRD13C | 0.762361 | 3 | 0 | 3 |
| CUL4B | ATG2B | 0.753774 | 3 | 0 | 3 |
| CUL4B | GPRC5B | 0.751474 | 3 | 0 | 3 |
| CUL4B | ARHGAP31 | 0.749569 | 3 | 0 | 3 |
| CUL4B | YAP1 | 0.742438 | 4 | 0 | 4 |
| CUL4B | MTMR10 | 0.738447 | 7 | 0 | 7 |
| CUL4B | EIF2D | 0.734042 | 3 | 0 | 3 |
| CUL4B | ZMYM6 | 0.733934 | 3 | 0 | 3 |
| CUL4B | BARD1 | 0.730671 | 3 | 0 | 3 |
| CUL4B | MLKL | 0.729325 | 3 | 0 | 3 |
| CUL4B | TMEM150C | 0.729138 | 3 | 0 | 3 |
| CUL4B | SENP8 | 0.728636 | 5 | 0 | 4 |
For details and further investigation, click here