| Full name: C-X-C motif chemokine ligand 14 | Alias Symbol: BRAK|NJAC|bolekine|Kec|MIP-2g|BMAC|KS1 | ||
| Type: protein-coding gene | Cytoband: 5q31.1 | ||
| Entrez ID: 9547 | HGNC ID: HGNC:10640 | Ensembl Gene: ENSG00000145824 | OMIM ID: 604186 |
| Drug and gene relationship at DGIdb | |||
CXCL14 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04062 | Chemokine signaling pathway |
Expression of CXCL14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CXCL14 | 9547 | 218002_s_at | -0.0270 | 0.9948 | |
| GSE20347 | CXCL14 | 9547 | 218002_s_at | 0.5729 | 0.3970 | |
| GSE23400 | CXCL14 | 9547 | 218002_s_at | 0.9785 | 0.0010 | |
| GSE26886 | CXCL14 | 9547 | 222484_s_at | 1.3550 | 0.0257 | |
| GSE29001 | CXCL14 | 9547 | 218002_s_at | 0.0981 | 0.9391 | |
| GSE38129 | CXCL14 | 9547 | 218002_s_at | 0.9448 | 0.0509 | |
| GSE45670 | CXCL14 | 9547 | 218002_s_at | 1.3367 | 0.0068 | |
| GSE53622 | CXCL14 | 9547 | 20798 | 0.6563 | 0.0142 | |
| GSE53624 | CXCL14 | 9547 | 20798 | 0.7034 | 0.0001 | |
| GSE63941 | CXCL14 | 9547 | 218002_s_at | -0.4064 | 0.8901 | |
| GSE77861 | CXCL14 | 9547 | 222484_s_at | 1.2567 | 0.1505 | |
| GSE97050 | CXCL14 | 9547 | A_33_P3590259 | 1.4782 | 0.1936 | |
| SRP007169 | CXCL14 | 9547 | RNAseq | 1.5736 | 0.0073 | |
| SRP008496 | CXCL14 | 9547 | RNAseq | 2.6012 | 0.0003 | |
| SRP064894 | CXCL14 | 9547 | RNAseq | 0.6401 | 0.1567 | |
| SRP133303 | CXCL14 | 9547 | RNAseq | 1.2142 | 0.1113 | |
| SRP159526 | CXCL14 | 9547 | RNAseq | 1.2895 | 0.1540 | |
| SRP193095 | CXCL14 | 9547 | RNAseq | 0.6953 | 0.0488 | |
| SRP219564 | CXCL14 | 9547 | RNAseq | -0.2741 | 0.7996 | |
| TCGA | CXCL14 | 9547 | RNAseq | 0.4451 | 0.0153 |
Upregulated datasets: 4; Downregulated datasets: 0.
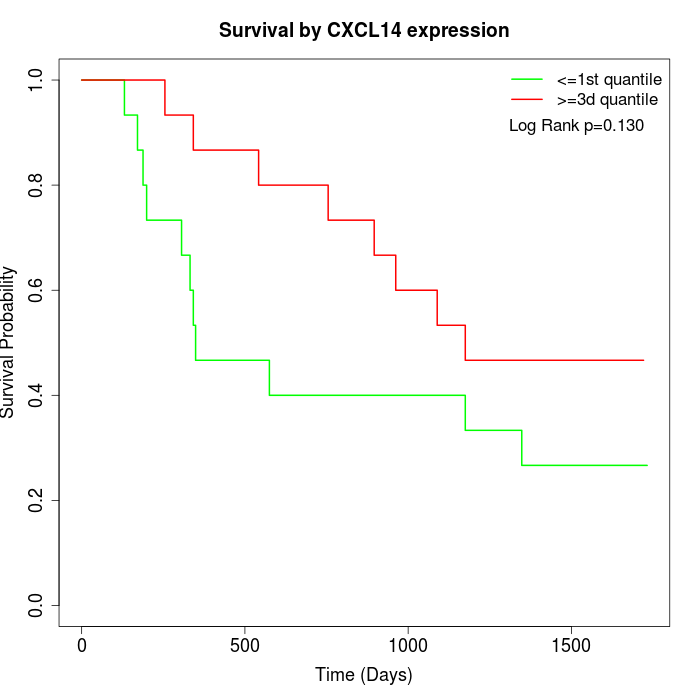
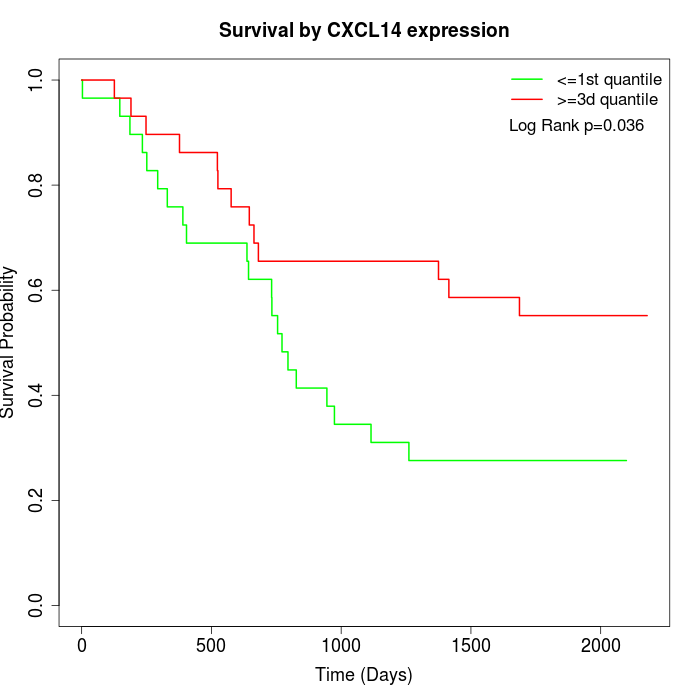
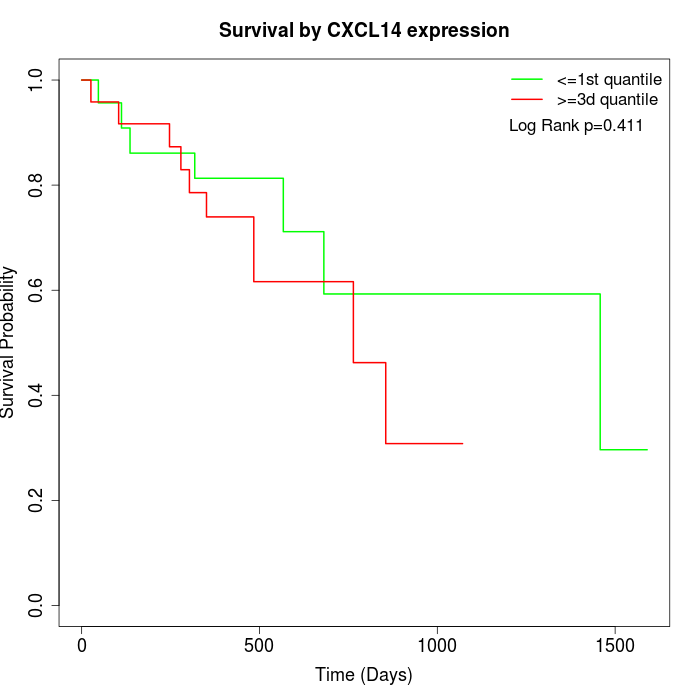
Survival by CXCL14 expression:
Note: Click image to view full size file.
Copy number change of CXCL14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CXCL14 | 9547 | 3 | 11 | 16 | |
| GSE20123 | CXCL14 | 9547 | 4 | 11 | 15 | |
| GSE43470 | CXCL14 | 9547 | 4 | 5 | 34 | |
| GSE46452 | CXCL14 | 9547 | 0 | 27 | 32 | |
| GSE47630 | CXCL14 | 9547 | 0 | 21 | 19 | |
| GSE54993 | CXCL14 | 9547 | 9 | 1 | 60 | |
| GSE54994 | CXCL14 | 9547 | 3 | 13 | 37 | |
| GSE60625 | CXCL14 | 9547 | 0 | 0 | 11 | |
| GSE74703 | CXCL14 | 9547 | 3 | 4 | 29 | |
| GSE74704 | CXCL14 | 9547 | 3 | 5 | 12 | |
| TCGA | CXCL14 | 9547 | 4 | 36 | 56 |
Total number of gains: 33; Total number of losses: 134; Total Number of normals: 321.
Somatic mutations of CXCL14:
Generating mutation plots.
Highly correlated genes for CXCL14:
Showing top 20/148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CXCL14 | ARSI | 0.689168 | 4 | 0 | 4 |
| CXCL14 | PRR34-AS1 | 0.680455 | 3 | 0 | 3 |
| CXCL14 | RELL2 | 0.669446 | 3 | 0 | 3 |
| CXCL14 | PCBP4 | 0.664022 | 3 | 0 | 3 |
| CXCL14 | BAHCC1 | 0.663429 | 3 | 0 | 3 |
| CXCL14 | CIC | 0.6533 | 3 | 0 | 3 |
| CXCL14 | SHANK3 | 0.645625 | 3 | 0 | 3 |
| CXCL14 | TENM2 | 0.645526 | 5 | 0 | 4 |
| CXCL14 | SMIM3 | 0.643828 | 3 | 0 | 3 |
| CXCL14 | SDK1 | 0.638992 | 3 | 0 | 3 |
| CXCL14 | OGFOD2 | 0.632679 | 3 | 0 | 3 |
| CXCL14 | CASC16 | 0.631408 | 3 | 0 | 3 |
| CXCL14 | MSN | 0.631296 | 4 | 0 | 3 |
| CXCL14 | SCARB1 | 0.630982 | 3 | 0 | 3 |
| CXCL14 | IGSF8 | 0.628951 | 3 | 0 | 3 |
| CXCL14 | HEXB | 0.628146 | 4 | 0 | 3 |
| CXCL14 | TRADD | 0.61445 | 3 | 0 | 3 |
| CXCL14 | WFDC5 | 0.612217 | 5 | 0 | 4 |
| CXCL14 | SLC25A44 | 0.609806 | 3 | 0 | 3 |
| CXCL14 | TNFRSF12A | 0.606072 | 4 | 0 | 3 |
For details and further investigation, click here