| Full name: PZP alpha-2-macroglobulin like | Alias Symbol: CPAMD6 | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 5858 | HGNC ID: HGNC:9750 | Ensembl Gene: ENSG00000126838 | OMIM ID: 176420 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PZP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PZP | 5858 | 207330_at | -0.2722 | 0.3680 | |
| GSE20347 | PZP | 5858 | 207330_at | 0.0756 | 0.4347 | |
| GSE23400 | PZP | 5858 | 207330_at | -0.0443 | 0.3232 | |
| GSE26886 | PZP | 5858 | 207330_at | 0.0296 | 0.8506 | |
| GSE29001 | PZP | 5858 | 207330_at | -0.0841 | 0.5620 | |
| GSE38129 | PZP | 5858 | 207330_at | -0.1914 | 0.0631 | |
| GSE45670 | PZP | 5858 | 207330_at | -0.1642 | 0.1516 | |
| GSE53622 | PZP | 5858 | 24486 | -0.7949 | 0.0000 | |
| GSE53624 | PZP | 5858 | 24486 | -0.8766 | 0.0000 | |
| GSE63941 | PZP | 5858 | 207330_at | -0.0450 | 0.7822 | |
| GSE77861 | PZP | 5858 | 207330_at | -0.1242 | 0.2693 | |
| GSE97050 | PZP | 5858 | A_23_P139682 | 0.0855 | 0.8528 | |
| SRP133303 | PZP | 5858 | RNAseq | -1.1995 | 0.0019 | |
| TCGA | PZP | 5858 | RNAseq | -0.5315 | 0.5432 |
Upregulated datasets: 0; Downregulated datasets: 1.
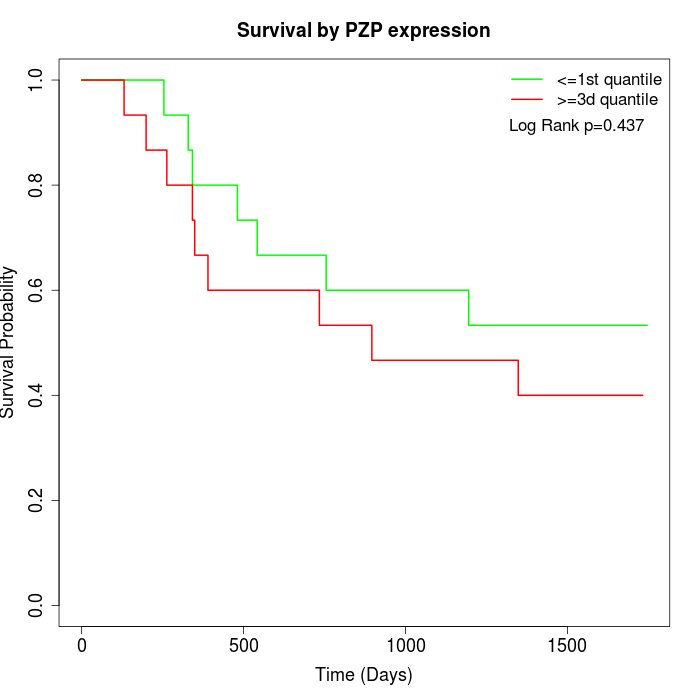
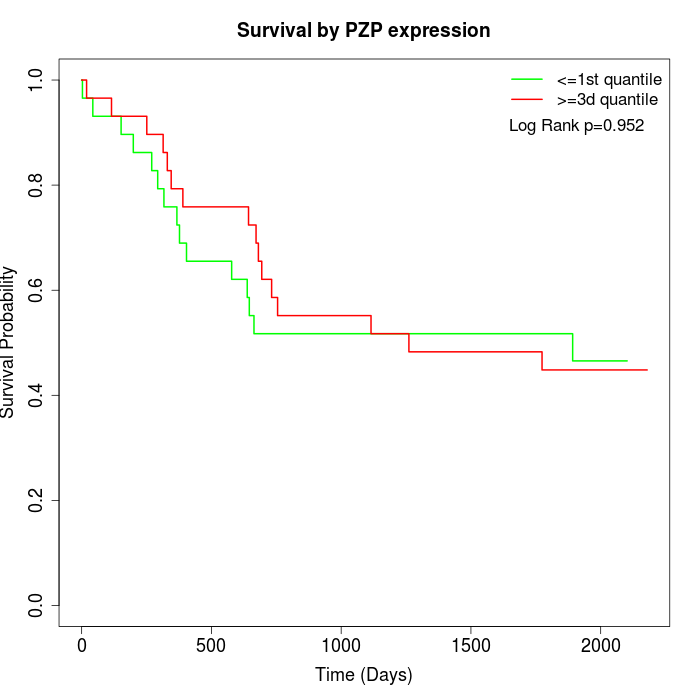
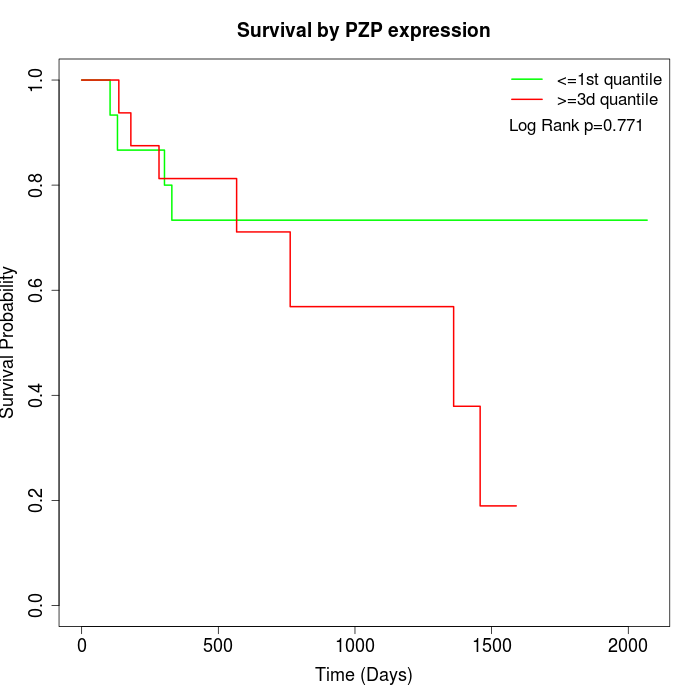
Survival by PZP expression:
Note: Click image to view full size file.
Copy number change of PZP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PZP | 5858 | 6 | 3 | 21 | |
| GSE20123 | PZP | 5858 | 6 | 3 | 21 | |
| GSE43470 | PZP | 5858 | 8 | 5 | 30 | |
| GSE46452 | PZP | 5858 | 10 | 1 | 48 | |
| GSE47630 | PZP | 5858 | 12 | 2 | 26 | |
| GSE54993 | PZP | 5858 | 1 | 10 | 59 | |
| GSE54994 | PZP | 5858 | 9 | 2 | 42 | |
| GSE60625 | PZP | 5858 | 0 | 1 | 10 | |
| GSE74703 | PZP | 5858 | 8 | 3 | 25 | |
| GSE74704 | PZP | 5858 | 4 | 2 | 14 | |
| TCGA | PZP | 5858 | 40 | 6 | 50 |
Total number of gains: 104; Total number of losses: 38; Total Number of normals: 346.
Somatic mutations of PZP:
Generating mutation plots.
Highly correlated genes for PZP:
Showing top 20/859 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PZP | KRT71 | 0.786814 | 3 | 0 | 3 |
| PZP | GNG11 | 0.74437 | 4 | 0 | 4 |
| PZP | ECSCR | 0.739482 | 5 | 0 | 5 |
| PZP | GNG2 | 0.737751 | 3 | 0 | 3 |
| PZP | TNFSF12 | 0.737733 | 3 | 0 | 3 |
| PZP | TCEAL5 | 0.736535 | 3 | 0 | 3 |
| PZP | TOM1L1 | 0.735349 | 3 | 0 | 3 |
| PZP | PODN | 0.728277 | 4 | 0 | 4 |
| PZP | CXCR5 | 0.728099 | 4 | 0 | 3 |
| PZP | C22orf31 | 0.727755 | 3 | 0 | 3 |
| PZP | GIMAP8 | 0.726855 | 4 | 0 | 4 |
| PZP | GIMAP5 | 0.724892 | 3 | 0 | 3 |
| PZP | ZNF619 | 0.724079 | 3 | 0 | 3 |
| PZP | STAC | 0.719761 | 3 | 0 | 3 |
| PZP | FRZB | 0.716828 | 3 | 0 | 3 |
| PZP | NR2F1 | 0.716563 | 3 | 0 | 3 |
| PZP | ARHGAP15 | 0.714393 | 3 | 0 | 3 |
| PZP | HIP1 | 0.705614 | 3 | 0 | 3 |
| PZP | PCDH18 | 0.704086 | 4 | 0 | 4 |
| PZP | SPON1 | 0.703711 | 4 | 0 | 4 |
For details and further investigation, click here