| Full name: DnaJ heat shock protein family (Hsp40) member B12 | Alias Symbol: DJ10|FLJ20027 | ||
| Type: protein-coding gene | Cytoband: 10q22.1 | ||
| Entrez ID: 54788 | HGNC ID: HGNC:14891 | Ensembl Gene: ENSG00000148719 | OMIM ID: 608376 |
| Drug and gene relationship at DGIdb | |||
Expression of DNAJB12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DNAJB12 | 54788 | 202866_at | -0.0825 | 0.8197 | |
| GSE20347 | DNAJB12 | 54788 | 202866_at | -0.2166 | 0.0050 | |
| GSE23400 | DNAJB12 | 54788 | 202866_at | -0.1440 | 0.0004 | |
| GSE26886 | DNAJB12 | 54788 | 202866_at | 0.1247 | 0.4277 | |
| GSE29001 | DNAJB12 | 54788 | 202866_at | -0.0906 | 0.6276 | |
| GSE38129 | DNAJB12 | 54788 | 202866_at | -0.0788 | 0.5160 | |
| GSE45670 | DNAJB12 | 54788 | 202866_at | -0.0107 | 0.9207 | |
| GSE53622 | DNAJB12 | 54788 | 144337 | -0.2015 | 0.0024 | |
| GSE53624 | DNAJB12 | 54788 | 144337 | 0.0707 | 0.1803 | |
| GSE63941 | DNAJB12 | 54788 | 202866_at | -0.2354 | 0.4739 | |
| GSE77861 | DNAJB12 | 54788 | 202866_at | -0.0942 | 0.5157 | |
| GSE97050 | DNAJB12 | 54788 | A_24_P244100 | 0.0103 | 0.9646 | |
| SRP007169 | DNAJB12 | 54788 | RNAseq | -0.4060 | 0.2451 | |
| SRP008496 | DNAJB12 | 54788 | RNAseq | -0.1190 | 0.5361 | |
| SRP064894 | DNAJB12 | 54788 | RNAseq | -0.1050 | 0.4912 | |
| SRP133303 | DNAJB12 | 54788 | RNAseq | 0.0383 | 0.7772 | |
| SRP159526 | DNAJB12 | 54788 | RNAseq | -0.3062 | 0.3081 | |
| SRP193095 | DNAJB12 | 54788 | RNAseq | -0.0463 | 0.4670 | |
| SRP219564 | DNAJB12 | 54788 | RNAseq | -0.2188 | 0.3397 | |
| TCGA | DNAJB12 | 54788 | RNAseq | -0.0669 | 0.1614 |
Upregulated datasets: 0; Downregulated datasets: 0.
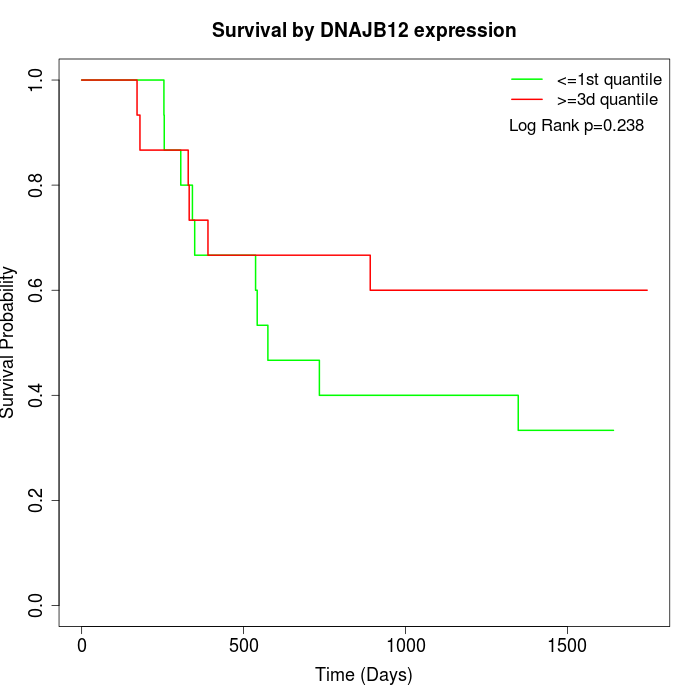
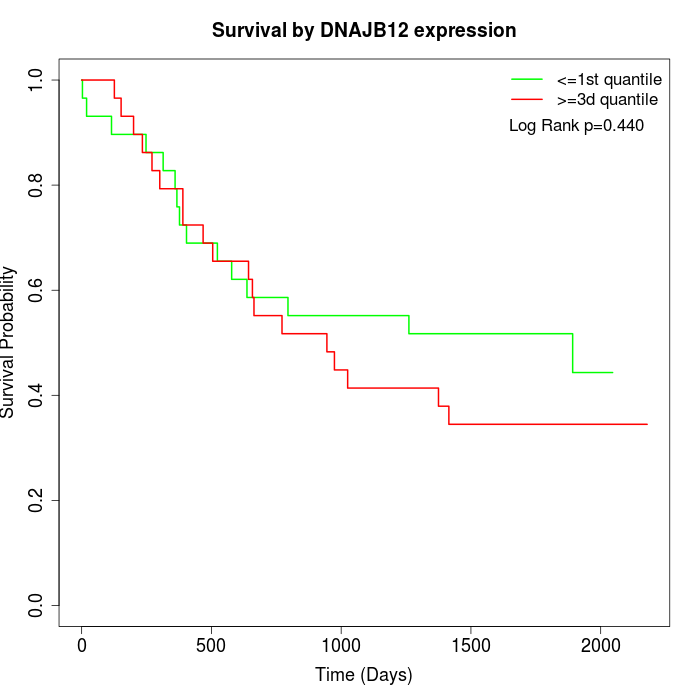
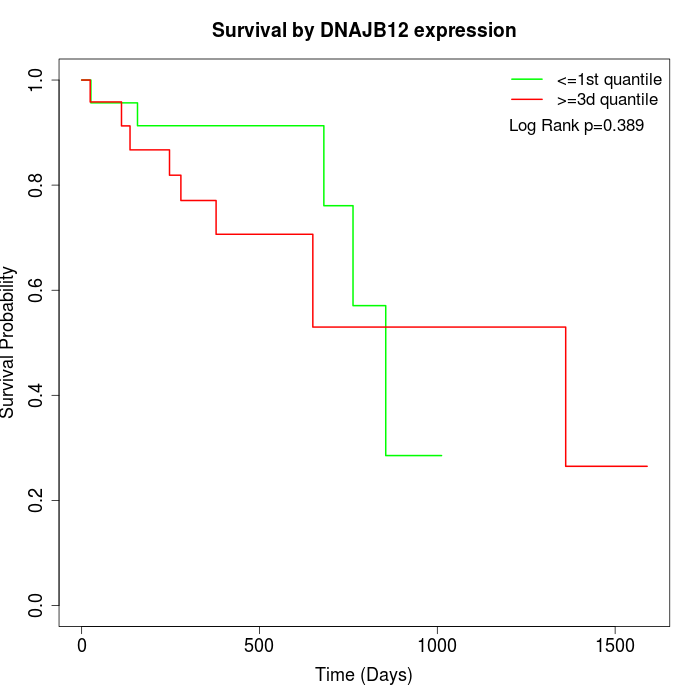
Survival by DNAJB12 expression:
Note: Click image to view full size file.
Copy number change of DNAJB12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DNAJB12 | 54788 | 3 | 4 | 23 | |
| GSE20123 | DNAJB12 | 54788 | 3 | 3 | 24 | |
| GSE43470 | DNAJB12 | 54788 | 1 | 8 | 34 | |
| GSE46452 | DNAJB12 | 54788 | 0 | 11 | 48 | |
| GSE47630 | DNAJB12 | 54788 | 2 | 14 | 24 | |
| GSE54993 | DNAJB12 | 54788 | 7 | 0 | 63 | |
| GSE54994 | DNAJB12 | 54788 | 2 | 11 | 40 | |
| GSE60625 | DNAJB12 | 54788 | 0 | 0 | 11 | |
| GSE74703 | DNAJB12 | 54788 | 1 | 5 | 30 | |
| GSE74704 | DNAJB12 | 54788 | 2 | 1 | 17 | |
| TCGA | DNAJB12 | 54788 | 11 | 23 | 62 |
Total number of gains: 32; Total number of losses: 80; Total Number of normals: 376.
Somatic mutations of DNAJB12:
Generating mutation plots.
Highly correlated genes for DNAJB12:
Showing top 20/379 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DNAJB12 | GPR132 | 0.783252 | 3 | 0 | 3 |
| DNAJB12 | RNF207 | 0.782919 | 3 | 0 | 3 |
| DNAJB12 | ZNF358 | 0.745219 | 3 | 0 | 3 |
| DNAJB12 | THRAP3 | 0.737202 | 3 | 0 | 3 |
| DNAJB12 | TSNARE1 | 0.731518 | 3 | 0 | 3 |
| DNAJB12 | CTDSP1 | 0.723075 | 5 | 0 | 5 |
| DNAJB12 | PITHD1 | 0.720235 | 3 | 0 | 3 |
| DNAJB12 | CXCR5 | 0.718026 | 3 | 0 | 3 |
| DNAJB12 | HDAC6 | 0.707342 | 3 | 0 | 3 |
| DNAJB12 | ABI1 | 0.7062 | 3 | 0 | 3 |
| DNAJB12 | VWA1 | 0.706081 | 3 | 0 | 3 |
| DNAJB12 | ZNF467 | 0.698744 | 3 | 0 | 3 |
| DNAJB12 | HIGD2A | 0.697926 | 3 | 0 | 3 |
| DNAJB12 | TRIM33 | 0.696714 | 4 | 0 | 4 |
| DNAJB12 | UBL5 | 0.696054 | 4 | 0 | 3 |
| DNAJB12 | CYFIP1 | 0.69513 | 3 | 0 | 3 |
| DNAJB12 | NLGN3 | 0.694271 | 3 | 0 | 3 |
| DNAJB12 | PRKAG1 | 0.692611 | 3 | 0 | 3 |
| DNAJB12 | LGI2 | 0.690921 | 3 | 0 | 3 |
| DNAJB12 | PRDM10 | 0.69071 | 3 | 0 | 3 |
For details and further investigation, click here