| Full name: ATPase Na+/K+ transporting subunit alpha 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1q23.2 | ||
| Entrez ID: 480 | HGNC ID: HGNC:14073 | Ensembl Gene: ENSG00000132681 | OMIM ID: 607321 |
| Drug and gene relationship at DGIdb | |||
ATP1A4 involved pathways:
Expression of ATP1A4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP1A4 | 480 | 1558612_a_at | -0.1440 | 0.5666 | |
| GSE26886 | ATP1A4 | 480 | 1558612_a_at | -0.1328 | 0.2113 | |
| GSE45670 | ATP1A4 | 480 | 1558612_a_at | 0.1193 | 0.2332 | |
| GSE53622 | ATP1A4 | 480 | 12536 | 0.3134 | 0.0001 | |
| GSE53624 | ATP1A4 | 480 | 12536 | 0.5227 | 0.0000 | |
| GSE63941 | ATP1A4 | 480 | 1558612_a_at | 0.1459 | 0.2551 | |
| GSE77861 | ATP1A4 | 480 | 1564241_at | -0.1344 | 0.2829 | |
| GSE97050 | ATP1A4 | 480 | A_24_P307626 | 0.1890 | 0.4562 | |
| TCGA | ATP1A4 | 480 | RNAseq | 1.6024 | 0.2137 |
Upregulated datasets: 0; Downregulated datasets: 0.
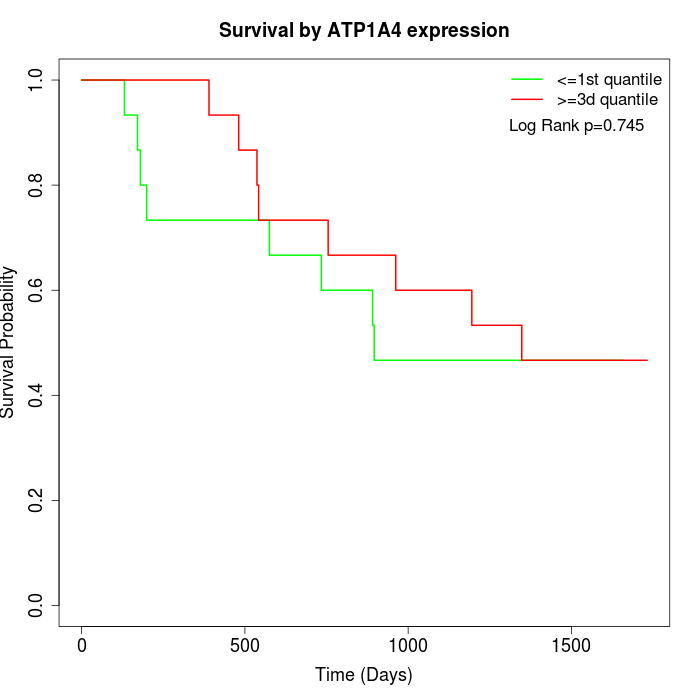
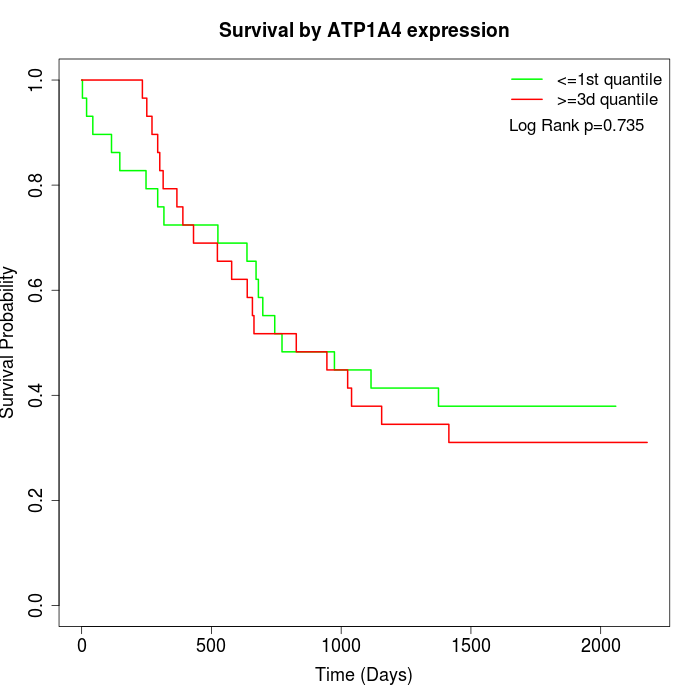
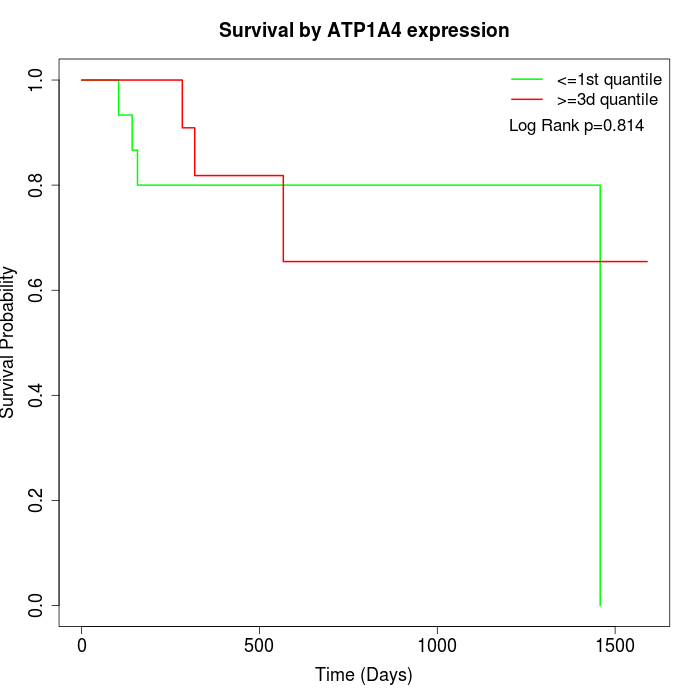
Survival by ATP1A4 expression:
Note: Click image to view full size file.
Copy number change of ATP1A4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP1A4 | 480 | 10 | 0 | 20 | |
| GSE20123 | ATP1A4 | 480 | 10 | 0 | 20 | |
| GSE43470 | ATP1A4 | 480 | 7 | 2 | 34 | |
| GSE46452 | ATP1A4 | 480 | 2 | 1 | 56 | |
| GSE47630 | ATP1A4 | 480 | 15 | 0 | 25 | |
| GSE54993 | ATP1A4 | 480 | 0 | 5 | 65 | |
| GSE54994 | ATP1A4 | 480 | 16 | 0 | 37 | |
| GSE60625 | ATP1A4 | 480 | 0 | 0 | 11 | |
| GSE74703 | ATP1A4 | 480 | 7 | 2 | 27 | |
| GSE74704 | ATP1A4 | 480 | 4 | 0 | 16 | |
| TCGA | ATP1A4 | 480 | 44 | 3 | 49 |
Total number of gains: 115; Total number of losses: 13; Total Number of normals: 360.
Somatic mutations of ATP1A4:
Generating mutation plots.
Highly correlated genes for ATP1A4:
Showing top 20/72 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP1A4 | GTPBP10 | 0.795585 | 3 | 0 | 3 |
| ATP1A4 | CXCR5 | 0.737098 | 3 | 0 | 3 |
| ATP1A4 | RNF207 | 0.736032 | 3 | 0 | 3 |
| ATP1A4 | KLF14 | 0.729709 | 3 | 0 | 3 |
| ATP1A4 | ZNF625 | 0.719226 | 3 | 0 | 3 |
| ATP1A4 | GPR37L1 | 0.714799 | 4 | 0 | 4 |
| ATP1A4 | SSNA1 | 0.712079 | 3 | 0 | 3 |
| ATP1A4 | CISD3 | 0.711157 | 3 | 0 | 3 |
| ATP1A4 | CIDEB | 0.704538 | 3 | 0 | 3 |
| ATP1A4 | HPCA | 0.70343 | 4 | 0 | 3 |
| ATP1A4 | MAPRE3 | 0.687485 | 3 | 0 | 3 |
| ATP1A4 | CISH | 0.682771 | 3 | 0 | 3 |
| ATP1A4 | BAGE | 0.678405 | 3 | 0 | 3 |
| ATP1A4 | NDUFA2 | 0.677005 | 3 | 0 | 3 |
| ATP1A4 | ITPRIP | 0.673465 | 3 | 0 | 3 |
| ATP1A4 | GSG1 | 0.668992 | 3 | 0 | 3 |
| ATP1A4 | LIPF | 0.668953 | 4 | 0 | 3 |
| ATP1A4 | PGLS | 0.664408 | 3 | 0 | 3 |
| ATP1A4 | ADAMTSL5 | 0.66371 | 3 | 0 | 3 |
| ATP1A4 | TPSD1 | 0.662638 | 3 | 0 | 3 |
For details and further investigation, click here