| Full name: NK2 homeobox 5 | Alias Symbol: CSX1|NKX2.5|NKX4-1 | ||
| Type: protein-coding gene | Cytoband: 5q34 | ||
| Entrez ID: 1482 | HGNC ID: HGNC:2488 | Ensembl Gene: ENSG00000183072 | OMIM ID: 600584 |
| Drug and gene relationship at DGIdb | |||
Expression of NKX2-5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NKX2-5 | 1482 | 206578_at | 0.2104 | 0.6675 | |
| GSE20347 | NKX2-5 | 1482 | 206578_at | 0.1049 | 0.1552 | |
| GSE23400 | NKX2-5 | 1482 | 206578_at | 0.0060 | 0.8779 | |
| GSE26886 | NKX2-5 | 1482 | 206578_at | 0.2785 | 0.0619 | |
| GSE29001 | NKX2-5 | 1482 | 206578_at | -0.0149 | 0.9237 | |
| GSE38129 | NKX2-5 | 1482 | 206578_at | 0.0750 | 0.2572 | |
| GSE45670 | NKX2-5 | 1482 | 206578_at | 0.1626 | 0.0611 | |
| GSE53622 | NKX2-5 | 1482 | 117132 | -0.1022 | 0.0275 | |
| GSE53624 | NKX2-5 | 1482 | 117132 | 0.1913 | 0.0223 | |
| GSE63941 | NKX2-5 | 1482 | 206578_at | 0.7088 | 0.1728 | |
| GSE77861 | NKX2-5 | 1482 | 206578_at | 0.0487 | 0.6964 | |
| GSE97050 | NKX2-5 | 1482 | A_33_P3299599 | 0.1392 | 0.5745 | |
| SRP219564 | NKX2-5 | 1482 | RNAseq | 6.9023 | 0.0000 | |
| TCGA | NKX2-5 | 1482 | RNAseq | 1.7703 | 0.0164 |
Upregulated datasets: 2; Downregulated datasets: 0.
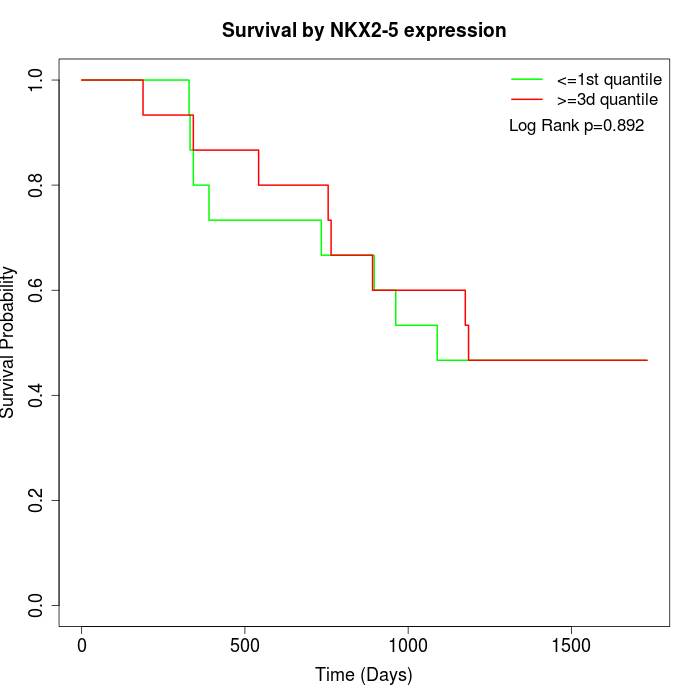
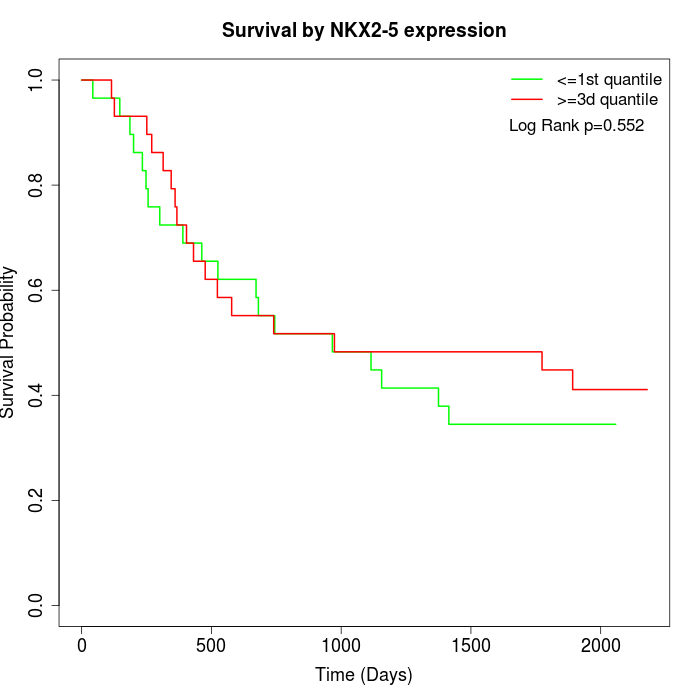
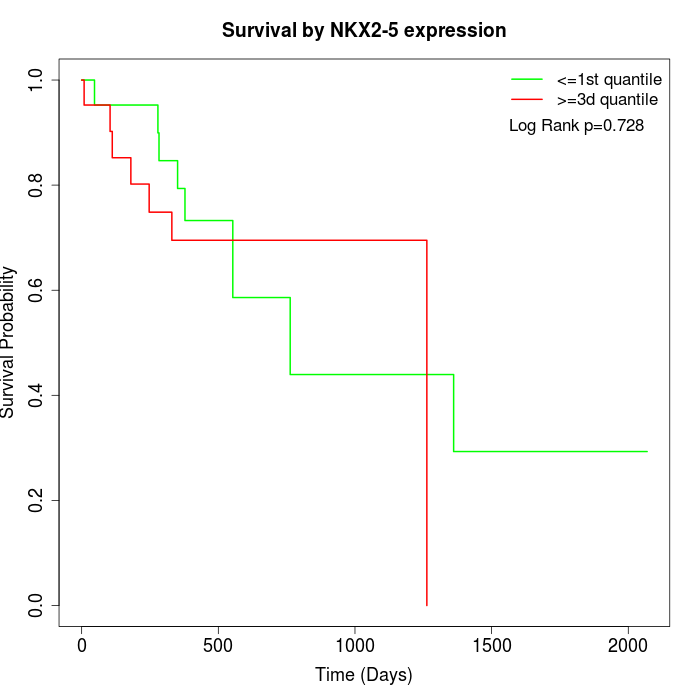
Survival by NKX2-5 expression:
Note: Click image to view full size file.
Copy number change of NKX2-5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NKX2-5 | 1482 | 2 | 13 | 15 | |
| GSE20123 | NKX2-5 | 1482 | 2 | 13 | 15 | |
| GSE43470 | NKX2-5 | 1482 | 2 | 10 | 31 | |
| GSE46452 | NKX2-5 | 1482 | 0 | 27 | 32 | |
| GSE47630 | NKX2-5 | 1482 | 0 | 20 | 20 | |
| GSE54993 | NKX2-5 | 1482 | 9 | 2 | 59 | |
| GSE54994 | NKX2-5 | 1482 | 2 | 17 | 34 | |
| GSE60625 | NKX2-5 | 1482 | 1 | 0 | 10 | |
| GSE74703 | NKX2-5 | 1482 | 2 | 7 | 27 | |
| GSE74704 | NKX2-5 | 1482 | 1 | 6 | 13 |
Total number of gains: 21; Total number of losses: 115; Total Number of normals: 256.
Somatic mutations of NKX2-5:
Generating mutation plots.
Highly correlated genes for NKX2-5:
Showing top 20/314 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NKX2-5 | CXCR5 | 0.815005 | 3 | 0 | 3 |
| NKX2-5 | PPP1R1B | 0.752684 | 3 | 0 | 3 |
| NKX2-5 | C12orf50 | 0.747691 | 3 | 0 | 3 |
| NKX2-5 | CRYBA1 | 0.738708 | 3 | 0 | 3 |
| NKX2-5 | SZT2 | 0.730647 | 3 | 0 | 3 |
| NKX2-5 | UNC5A | 0.72945 | 3 | 0 | 3 |
| NKX2-5 | DLL4 | 0.715622 | 3 | 0 | 3 |
| NKX2-5 | ITGB3 | 0.710581 | 3 | 0 | 3 |
| NKX2-5 | LRRC36 | 0.709104 | 3 | 0 | 3 |
| NKX2-5 | ERC1 | 0.701259 | 3 | 0 | 3 |
| NKX2-5 | KHDC1 | 0.698574 | 3 | 0 | 3 |
| NKX2-5 | TMSB15B | 0.695703 | 3 | 0 | 3 |
| NKX2-5 | STAC | 0.694284 | 3 | 0 | 3 |
| NKX2-5 | FAM167B | 0.691738 | 4 | 0 | 3 |
| NKX2-5 | KRTAP5-8 | 0.687734 | 3 | 0 | 3 |
| NKX2-5 | PCBP3 | 0.682371 | 4 | 0 | 3 |
| NKX2-5 | FIBCD1 | 0.678866 | 5 | 0 | 4 |
| NKX2-5 | LAMTOR1 | 0.67759 | 3 | 0 | 3 |
| NKX2-5 | COLEC11 | 0.676152 | 3 | 0 | 3 |
| NKX2-5 | PDGFB | 0.666052 | 5 | 0 | 5 |
For details and further investigation, click here