| Full name: cytochrome P450 family 4 subfamily Z member 1 | Alias Symbol: CYP4A20 | ||
| Type: protein-coding gene | Cytoband: 1p33 | ||
| Entrez ID: 199974 | HGNC ID: HGNC:20583 | Ensembl Gene: ENSG00000186160 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of CYP4Z1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CYP4Z1 | 199974 | 237395_at | 0.3139 | 0.6876 | |
| GSE26886 | CYP4Z1 | 199974 | 237395_at | 0.2017 | 0.5307 | |
| GSE45670 | CYP4Z1 | 199974 | 237395_at | 0.0712 | 0.8549 | |
| GSE53622 | CYP4Z1 | 199974 | 28685 | -0.4075 | 0.0433 | |
| GSE53624 | CYP4Z1 | 199974 | 28685 | -1.3838 | 0.0000 | |
| GSE63941 | CYP4Z1 | 199974 | 237395_at | 0.1531 | 0.4548 | |
| GSE77861 | CYP4Z1 | 199974 | 237395_at | -0.1670 | 0.0740 | |
| GSE97050 | CYP4Z1 | A_21_P0010756 | 0.1732 | 0.3810 | ||
| SRP133303 | CYP4Z1 | 199974 | RNAseq | -1.6239 | 0.0000 | |
| TCGA | CYP4Z1 | 199974 | RNAseq | 0.5764 | 0.4602 |
Upregulated datasets: 0; Downregulated datasets: 2.
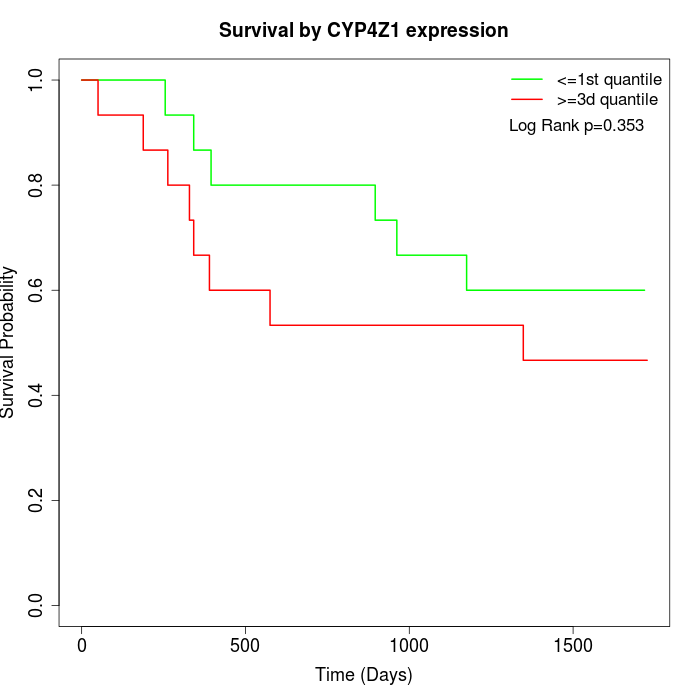
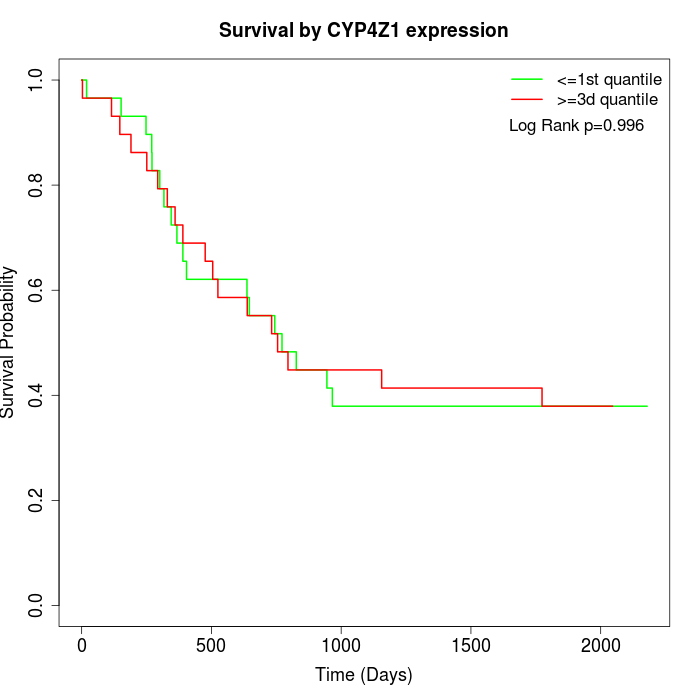
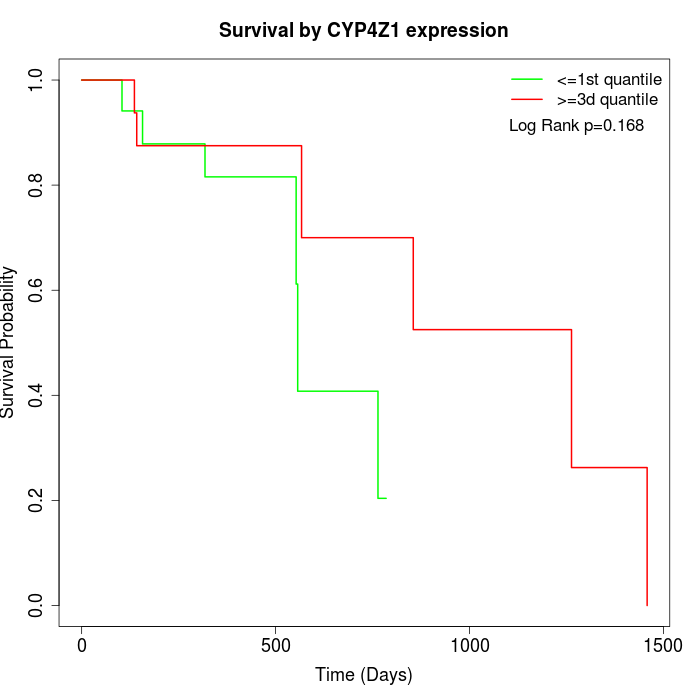
Survival by CYP4Z1 expression:
Note: Click image to view full size file.
Copy number change of CYP4Z1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CYP4Z1 | 199974 | 4 | 3 | 23 | |
| GSE20123 | CYP4Z1 | 199974 | 4 | 2 | 24 | |
| GSE43470 | CYP4Z1 | 199974 | 5 | 1 | 37 | |
| GSE46452 | CYP4Z1 | 199974 | 2 | 1 | 56 | |
| GSE47630 | CYP4Z1 | 199974 | 8 | 4 | 28 | |
| GSE54993 | CYP4Z1 | 199974 | 0 | 1 | 69 | |
| GSE54994 | CYP4Z1 | 199974 | 11 | 2 | 40 | |
| GSE60625 | CYP4Z1 | 199974 | 0 | 0 | 11 | |
| GSE74703 | CYP4Z1 | 199974 | 4 | 1 | 31 | |
| GSE74704 | CYP4Z1 | 199974 | 2 | 0 | 18 | |
| TCGA | CYP4Z1 | 199974 | 10 | 17 | 69 |
Total number of gains: 50; Total number of losses: 32; Total Number of normals: 406.
Somatic mutations of CYP4Z1:
Generating mutation plots.
Highly correlated genes for CYP4Z1:
Showing top 20/35 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CYP4Z1 | FBLN7 | 0.702324 | 3 | 0 | 3 |
| CYP4Z1 | VASH2 | 0.698589 | 3 | 0 | 3 |
| CYP4Z1 | GALK1 | 0.678175 | 3 | 0 | 3 |
| CYP4Z1 | GMEB1 | 0.657117 | 3 | 0 | 3 |
| CYP4Z1 | ZDHHC15 | 0.6513 | 3 | 0 | 3 |
| CYP4Z1 | DAPK2 | 0.64653 | 4 | 0 | 3 |
| CYP4Z1 | SYCP3 | 0.642116 | 4 | 0 | 3 |
| CYP4Z1 | TBX19 | 0.63495 | 3 | 0 | 3 |
| CYP4Z1 | C12orf54 | 0.634592 | 4 | 0 | 3 |
| CYP4Z1 | LRCH4 | 0.633544 | 4 | 0 | 4 |
| CYP4Z1 | PHKA2 | 0.624079 | 4 | 0 | 3 |
| CYP4Z1 | TTC21A | 0.615836 | 4 | 0 | 3 |
| CYP4Z1 | CAPN13 | 0.610705 | 5 | 0 | 3 |
| CYP4Z1 | VILL | 0.598073 | 5 | 0 | 3 |
| CYP4Z1 | CD8B | 0.596637 | 4 | 0 | 3 |
| CYP4Z1 | GNLY | 0.592123 | 3 | 0 | 3 |
| CYP4Z1 | UBXN11 | 0.584191 | 3 | 0 | 3 |
| CYP4Z1 | LINC00525 | 0.579009 | 4 | 0 | 3 |
| CYP4Z1 | LINC01222 | 0.565364 | 3 | 0 | 3 |
| CYP4Z1 | ABCB1 | 0.564941 | 5 | 0 | 4 |
For details and further investigation, click here