| Full name: phosphorylase kinase regulatory subunit alpha 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp22.13 | ||
| Entrez ID: 5256 | HGNC ID: HGNC:8926 | Ensembl Gene: ENSG00000044446 | OMIM ID: 300798 |
| Drug and gene relationship at DGIdb | |||
PHKA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway |
Expression of PHKA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PHKA2 | 5256 | 209439_s_at | 0.2658 | 0.6118 | |
| GSE20347 | PHKA2 | 5256 | 209439_s_at | 0.1046 | 0.3907 | |
| GSE23400 | PHKA2 | 5256 | 209439_s_at | 0.0110 | 0.8464 | |
| GSE26886 | PHKA2 | 5256 | 209439_s_at | 0.2587 | 0.3108 | |
| GSE29001 | PHKA2 | 5256 | 209439_s_at | 0.0918 | 0.6392 | |
| GSE38129 | PHKA2 | 5256 | 209439_s_at | 0.0155 | 0.8841 | |
| GSE45670 | PHKA2 | 5256 | 209439_s_at | 0.0916 | 0.6546 | |
| GSE53622 | PHKA2 | 5256 | 25049 | -0.2549 | 0.0029 | |
| GSE53624 | PHKA2 | 5256 | 122480 | 0.2004 | 0.0170 | |
| GSE63941 | PHKA2 | 5256 | 209439_s_at | 0.2433 | 0.6877 | |
| GSE77861 | PHKA2 | 5256 | 209439_s_at | 0.0136 | 0.9568 | |
| GSE97050 | PHKA2 | 5256 | A_23_P159671 | 0.0475 | 0.8219 | |
| SRP007169 | PHKA2 | 5256 | RNAseq | 0.5232 | 0.2776 | |
| SRP008496 | PHKA2 | 5256 | RNAseq | 0.8277 | 0.0167 | |
| SRP064894 | PHKA2 | 5256 | RNAseq | 0.1520 | 0.3006 | |
| SRP133303 | PHKA2 | 5256 | RNAseq | 0.1137 | 0.7031 | |
| SRP159526 | PHKA2 | 5256 | RNAseq | 0.2635 | 0.4338 | |
| SRP193095 | PHKA2 | 5256 | RNAseq | 0.0668 | 0.5916 | |
| SRP219564 | PHKA2 | 5256 | RNAseq | 0.2901 | 0.4419 | |
| TCGA | PHKA2 | 5256 | RNAseq | -0.1374 | 0.0337 |
Upregulated datasets: 0; Downregulated datasets: 0.
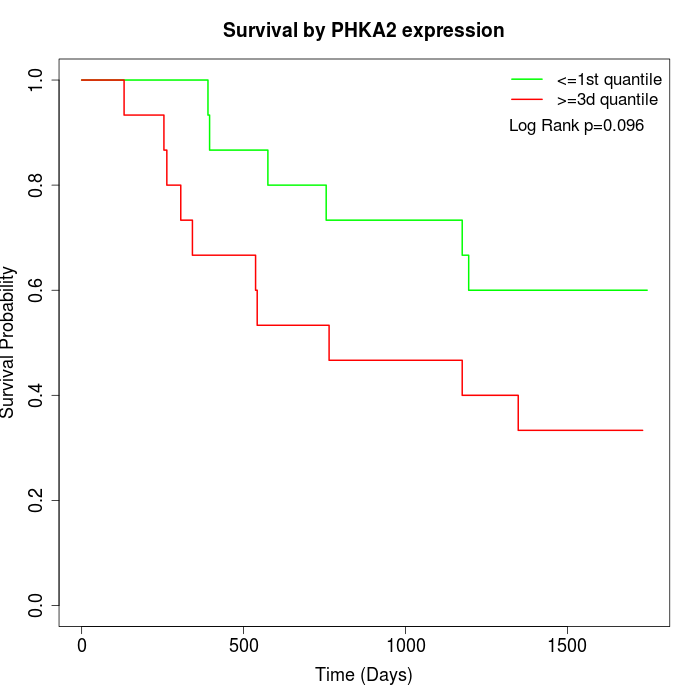
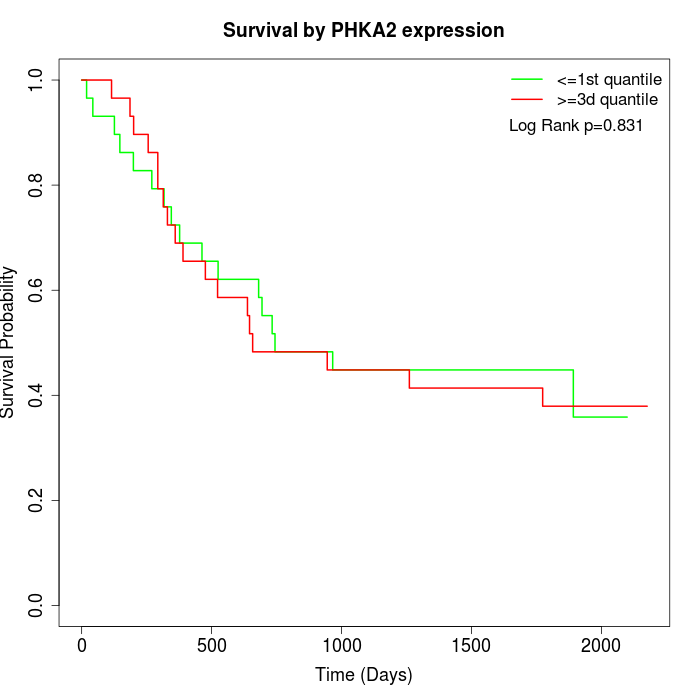
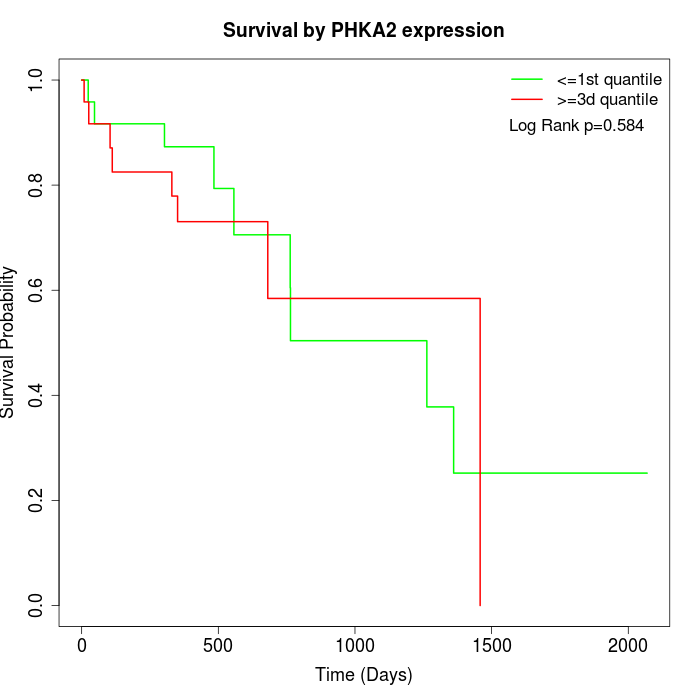
Survival by PHKA2 expression:
Note: Click image to view full size file.
Copy number change of PHKA2:
No record found for this gene.
Somatic mutations of PHKA2:
Generating mutation plots.
Highly correlated genes for PHKA2:
Showing top 20/67 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PHKA2 | BEND3 | 0.734259 | 3 | 0 | 3 |
| PHKA2 | PRDM13 | 0.729755 | 3 | 0 | 3 |
| PHKA2 | FAM24B | 0.721905 | 4 | 0 | 4 |
| PHKA2 | USP27X | 0.710367 | 4 | 0 | 3 |
| PHKA2 | ZBTB45 | 0.704658 | 3 | 0 | 3 |
| PHKA2 | ITGA2B | 0.695108 | 4 | 0 | 3 |
| PHKA2 | BATF2 | 0.692774 | 3 | 0 | 3 |
| PHKA2 | CCDC130 | 0.687456 | 4 | 0 | 3 |
| PHKA2 | ATF7IP2 | 0.679441 | 4 | 0 | 3 |
| PHKA2 | PRAME | 0.65711 | 3 | 0 | 3 |
| PHKA2 | MBD6 | 0.656521 | 4 | 0 | 3 |
| PHKA2 | GRIA2 | 0.644749 | 3 | 0 | 3 |
| PHKA2 | TRIM2 | 0.641936 | 3 | 0 | 3 |
| PHKA2 | CYP4Z1 | 0.624079 | 4 | 0 | 3 |
| PHKA2 | GNMT | 0.621637 | 4 | 0 | 3 |
| PHKA2 | PLEKHS1 | 0.617437 | 4 | 0 | 3 |
| PHKA2 | ZNF559 | 0.617054 | 4 | 0 | 3 |
| PHKA2 | PARS2 | 0.614569 | 4 | 0 | 3 |
| PHKA2 | LHX4-AS1 | 0.605781 | 3 | 0 | 3 |
| PHKA2 | E2F1 | 0.603846 | 5 | 0 | 3 |
For details and further investigation, click here