| Full name: decorin | Alias Symbol: DSPG2|SLRR1B | ||
| Type: protein-coding gene | Cytoband: 12q21.33 | ||
| Entrez ID: 1634 | HGNC ID: HGNC:2705 | Ensembl Gene: ENSG00000011465 | OMIM ID: 125255 |
| Drug and gene relationship at DGIdb | |||
DCN involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04350 | TGF-beta signaling pathway | |
| hsa05205 | Proteoglycans in cancer |
Expression of DCN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DCN | 1634 | 201893_x_at | -0.9382 | 0.4573 | |
| GSE20347 | DCN | 1634 | 201893_x_at | 0.5706 | 0.1214 | |
| GSE23400 | DCN | 1634 | 201893_x_at | -0.2740 | 0.1552 | |
| GSE26886 | DCN | 1634 | 201893_x_at | 0.5425 | 0.4122 | |
| GSE29001 | DCN | 1634 | 201893_x_at | -0.3240 | 0.4710 | |
| GSE38129 | DCN | 1634 | 201893_x_at | -0.0132 | 0.9815 | |
| GSE45670 | DCN | 1634 | 201893_x_at | -2.3586 | 0.0000 | |
| GSE53622 | DCN | 1634 | 65224 | -0.2670 | 0.1043 | |
| GSE53624 | DCN | 1634 | 65224 | 0.0755 | 0.6227 | |
| GSE63941 | DCN | 1634 | 211813_x_at | -8.6060 | 0.0000 | |
| GSE77861 | DCN | 1634 | 201893_x_at | 0.9727 | 0.0644 | |
| GSE97050 | DCN | 1634 | A_23_P64873 | -0.3887 | 0.5336 | |
| SRP007169 | DCN | 1634 | RNAseq | 1.8516 | 0.0009 | |
| SRP008496 | DCN | 1634 | RNAseq | 2.5073 | 0.0000 | |
| SRP064894 | DCN | 1634 | RNAseq | -0.3407 | 0.4092 | |
| SRP133303 | DCN | 1634 | RNAseq | -0.1260 | 0.7325 | |
| SRP159526 | DCN | 1634 | RNAseq | -0.6071 | 0.2068 | |
| SRP193095 | DCN | 1634 | RNAseq | 1.5726 | 0.0000 | |
| SRP219564 | DCN | 1634 | RNAseq | -0.0455 | 0.9576 | |
| TCGA | DCN | 1634 | RNAseq | -0.3534 | 0.0005 |
Upregulated datasets: 3; Downregulated datasets: 2.
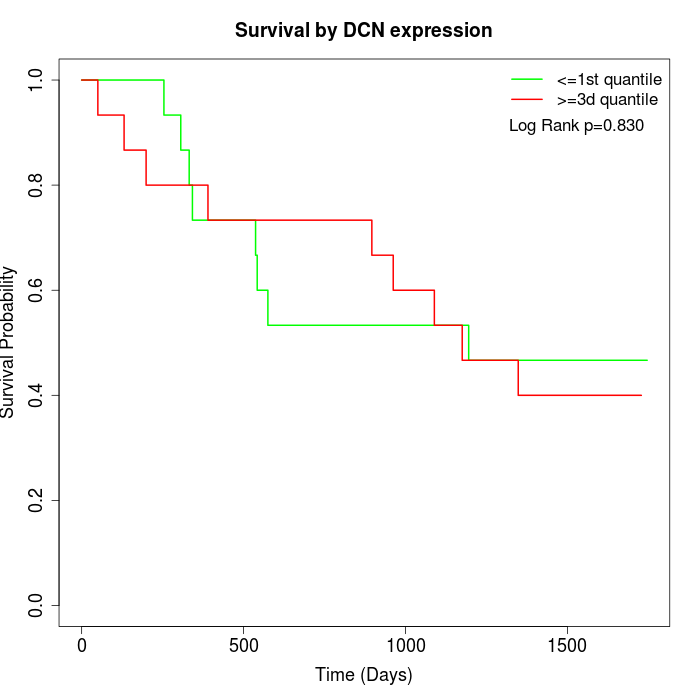
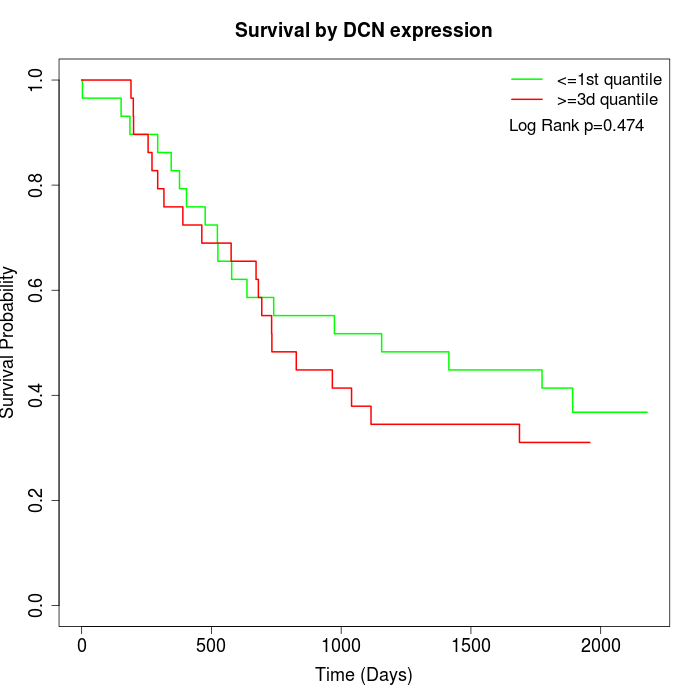
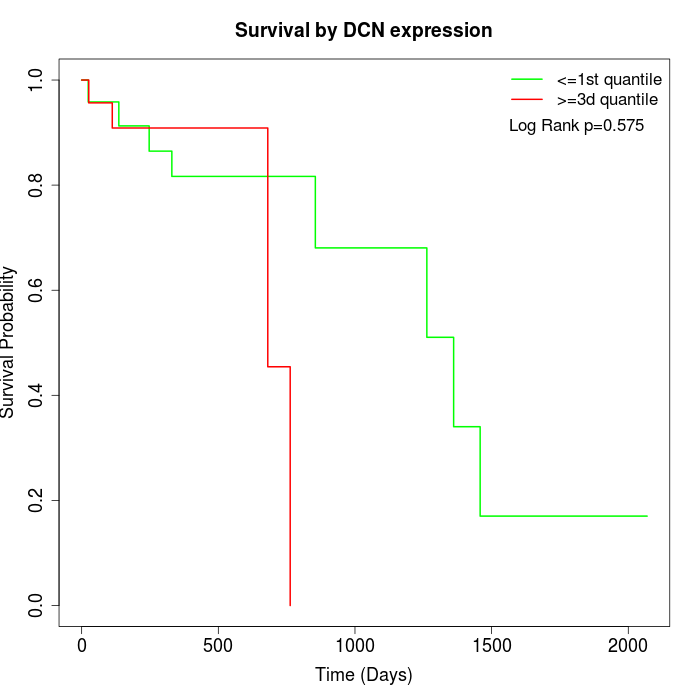
Survival by DCN expression:
Note: Click image to view full size file.
Copy number change of DCN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DCN | 1634 | 3 | 2 | 25 | |
| GSE20123 | DCN | 1634 | 3 | 2 | 25 | |
| GSE43470 | DCN | 1634 | 3 | 0 | 40 | |
| GSE46452 | DCN | 1634 | 9 | 1 | 49 | |
| GSE47630 | DCN | 1634 | 10 | 1 | 29 | |
| GSE54993 | DCN | 1634 | 0 | 6 | 64 | |
| GSE54994 | DCN | 1634 | 6 | 1 | 46 | |
| GSE60625 | DCN | 1634 | 0 | 0 | 11 | |
| GSE74703 | DCN | 1634 | 3 | 0 | 33 | |
| GSE74704 | DCN | 1634 | 1 | 2 | 17 | |
| TCGA | DCN | 1634 | 17 | 12 | 67 |
Total number of gains: 55; Total number of losses: 27; Total Number of normals: 406.
Somatic mutations of DCN:
Generating mutation plots.
Highly correlated genes for DCN:
Showing top 20/867 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DCN | OLFML3 | 0.801433 | 12 | 0 | 12 |
| DCN | CNRIP1 | 0.797759 | 9 | 0 | 9 |
| DCN | PMP22 | 0.775798 | 11 | 0 | 11 |
| DCN | FILIP1L | 0.766099 | 12 | 0 | 12 |
| DCN | DDR2 | 0.76347 | 12 | 0 | 11 |
| DCN | CALD1 | 0.756081 | 12 | 0 | 12 |
| DCN | ANXA6 | 0.755567 | 10 | 0 | 10 |
| DCN | ZNF583 | 0.751851 | 3 | 0 | 3 |
| DCN | FBN1 | 0.750537 | 11 | 0 | 10 |
| DCN | CCDC80 | 0.750335 | 8 | 0 | 7 |
| DCN | FBLN5 | 0.742946 | 11 | 0 | 10 |
| DCN | RARRES2 | 0.742378 | 12 | 0 | 12 |
| DCN | LAMA4 | 0.740726 | 11 | 0 | 11 |
| DCN | MRGPRF | 0.739749 | 8 | 0 | 7 |
| DCN | COLEC12 | 0.738649 | 11 | 0 | 9 |
| DCN | SFRP2 | 0.735101 | 7 | 0 | 6 |
| DCN | TEK | 0.733092 | 5 | 0 | 5 |
| DCN | OLFML1 | 0.731601 | 13 | 0 | 12 |
| DCN | PABPC5 | 0.726477 | 6 | 0 | 6 |
| DCN | TIMP2 | 0.725156 | 9 | 0 | 7 |
For details and further investigation, click here