| Full name: discoidin domain receptor tyrosine kinase 2 | Alias Symbol: TKT | ||
| Type: protein-coding gene | Cytoband: 1q23.3 | ||
| Entrez ID: 4921 | HGNC ID: HGNC:2731 | Ensembl Gene: ENSG00000162733 | OMIM ID: 191311 |
| Drug and gene relationship at DGIdb | |||
Expression of DDR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DDR2 | 4921 | 225442_at | -1.2300 | 0.3021 | |
| GSE20347 | DDR2 | 4921 | 205168_at | 0.2436 | 0.0791 | |
| GSE23400 | DDR2 | 4921 | 205168_at | -0.2805 | 0.0210 | |
| GSE26886 | DDR2 | 4921 | 227561_at | 0.4843 | 0.1111 | |
| GSE29001 | DDR2 | 4921 | 205168_at | 0.1842 | 0.6789 | |
| GSE38129 | DDR2 | 4921 | 205168_at | -0.3510 | 0.2930 | |
| GSE45670 | DDR2 | 4921 | 225442_at | -2.8198 | 0.0000 | |
| GSE53622 | DDR2 | 4921 | 7217 | -0.5845 | 0.0004 | |
| GSE53624 | DDR2 | 4921 | 7217 | -0.4631 | 0.0006 | |
| GSE63941 | DDR2 | 4921 | 227561_at | -5.9765 | 0.0000 | |
| GSE77861 | DDR2 | 4921 | 205168_at | 0.2100 | 0.1604 | |
| GSE97050 | DDR2 | 4921 | A_23_P452 | -0.7490 | 0.1057 | |
| SRP007169 | DDR2 | 4921 | RNAseq | 3.2607 | 0.0000 | |
| SRP008496 | DDR2 | 4921 | RNAseq | 3.9421 | 0.0000 | |
| SRP064894 | DDR2 | 4921 | RNAseq | 0.0092 | 0.9787 | |
| SRP133303 | DDR2 | 4921 | RNAseq | -0.1798 | 0.6313 | |
| SRP159526 | DDR2 | 4921 | RNAseq | 0.0597 | 0.8536 | |
| SRP193095 | DDR2 | 4921 | RNAseq | 1.4733 | 0.0000 | |
| SRP219564 | DDR2 | 4921 | RNAseq | -0.2589 | 0.7754 | |
| TCGA | DDR2 | 4921 | RNAseq | -0.6530 | 0.0007 |
Upregulated datasets: 3; Downregulated datasets: 2.
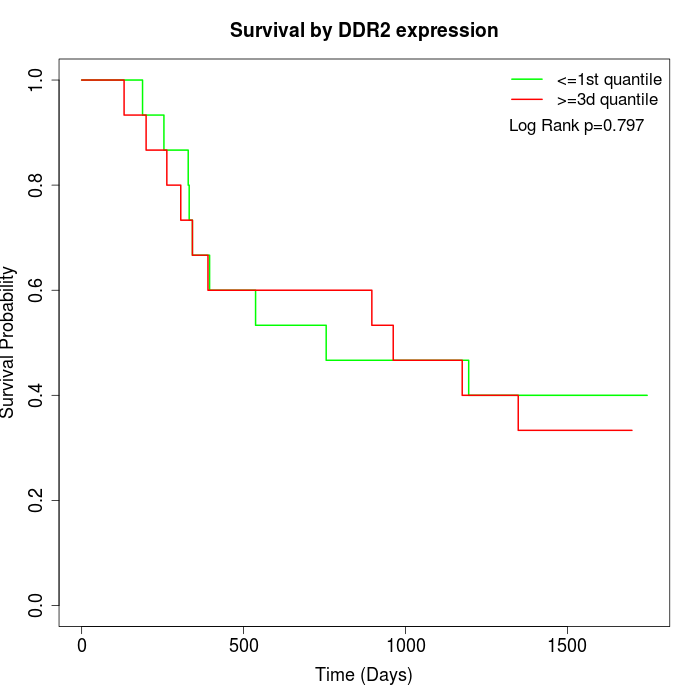
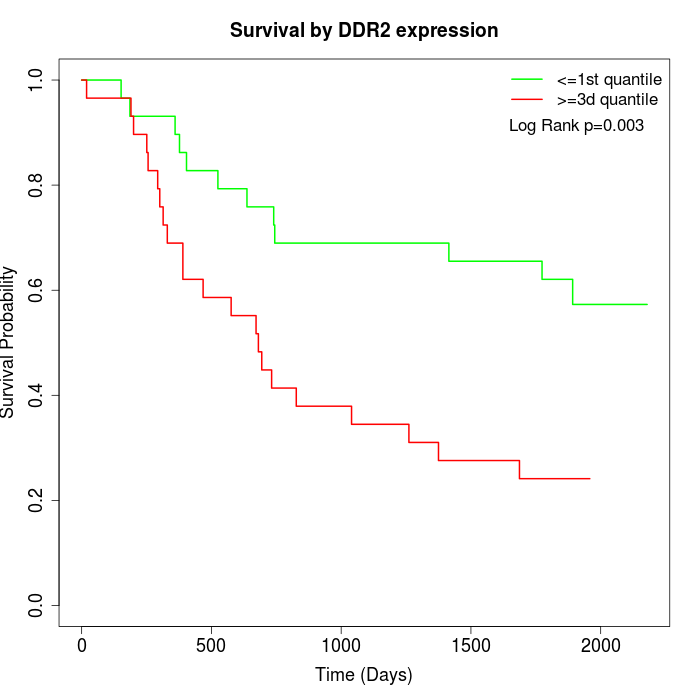
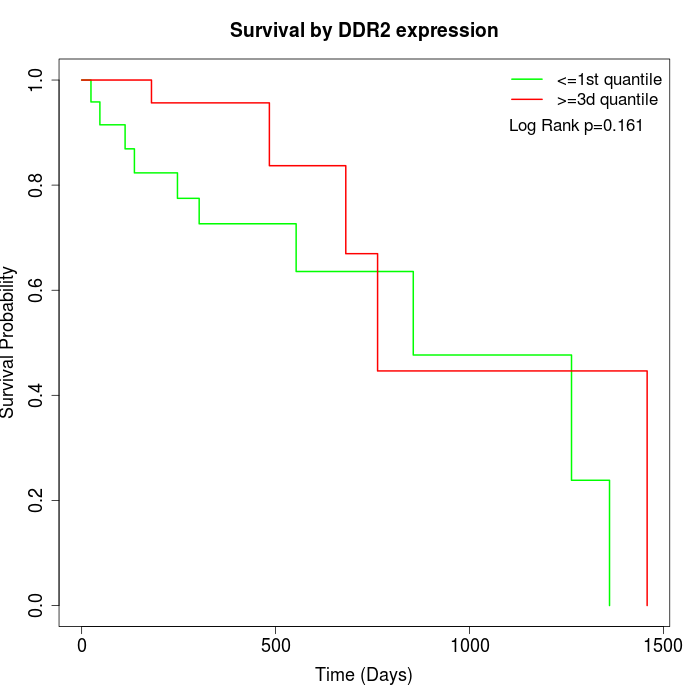
Survival by DDR2 expression:
Note: Click image to view full size file.
Copy number change of DDR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DDR2 | 4921 | 10 | 0 | 20 | |
| GSE20123 | DDR2 | 4921 | 10 | 0 | 20 | |
| GSE43470 | DDR2 | 4921 | 7 | 2 | 34 | |
| GSE46452 | DDR2 | 4921 | 2 | 1 | 56 | |
| GSE47630 | DDR2 | 4921 | 14 | 0 | 26 | |
| GSE54993 | DDR2 | 4921 | 0 | 5 | 65 | |
| GSE54994 | DDR2 | 4921 | 16 | 0 | 37 | |
| GSE60625 | DDR2 | 4921 | 0 | 0 | 11 | |
| GSE74703 | DDR2 | 4921 | 7 | 2 | 27 | |
| GSE74704 | DDR2 | 4921 | 4 | 0 | 16 | |
| TCGA | DDR2 | 4921 | 42 | 4 | 50 |
Total number of gains: 112; Total number of losses: 14; Total Number of normals: 362.
Somatic mutations of DDR2:
Generating mutation plots.
Highly correlated genes for DDR2:
Showing top 20/1259 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DDR2 | FIBIN | 0.872522 | 5 | 0 | 5 |
| DDR2 | LINC01279 | 0.861078 | 4 | 0 | 4 |
| DDR2 | MSRB3 | 0.858602 | 7 | 0 | 7 |
| DDR2 | RUNX1T1 | 0.848652 | 8 | 0 | 8 |
| DDR2 | MAGI2-AS3 | 0.844868 | 6 | 0 | 6 |
| DDR2 | CNRIP1 | 0.822743 | 9 | 0 | 9 |
| DDR2 | DNM3OS | 0.822231 | 4 | 0 | 4 |
| DDR2 | MRVI1 | 0.81864 | 8 | 0 | 8 |
| DDR2 | ZEB1 | 0.815514 | 3 | 0 | 3 |
| DDR2 | DIXDC1 | 0.814352 | 9 | 0 | 8 |
| DDR2 | ZFAND5 | 0.812928 | 3 | 0 | 3 |
| DDR2 | JAM3 | 0.803294 | 13 | 0 | 13 |
| DDR2 | TMEM47 | 0.801377 | 12 | 0 | 12 |
| DDR2 | ADARB1 | 0.796166 | 7 | 0 | 7 |
| DDR2 | ACTA2 | 0.792314 | 12 | 0 | 12 |
| DDR2 | MGARP | 0.791103 | 3 | 0 | 3 |
| DDR2 | PARVA | 0.787775 | 10 | 0 | 10 |
| DDR2 | FILIP1L | 0.787498 | 12 | 0 | 12 |
| DDR2 | SGCD | 0.78691 | 9 | 0 | 8 |
| DDR2 | TAGLN | 0.783119 | 11 | 0 | 11 |
For details and further investigation, click here