| Full name: caldesmon 1 | Alias Symbol: CDM|H-CAD|L-CAD | ||
| Type: protein-coding gene | Cytoband: 7q33 | ||
| Entrez ID: 800 | HGNC ID: HGNC:1441 | Ensembl Gene: ENSG00000122786 | OMIM ID: 114213 |
| Drug and gene relationship at DGIdb | |||
CALD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04270 | Vascular smooth muscle contraction |
Expression of CALD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CALD1 | 800 | 212077_at | -0.5345 | 0.4214 | |
| GSE20347 | CALD1 | 800 | 212077_at | 0.3761 | 0.3447 | |
| GSE23400 | CALD1 | 800 | 212077_at | 0.0246 | 0.9031 | |
| GSE26886 | CALD1 | 800 | 212077_at | 1.5210 | 0.0007 | |
| GSE29001 | CALD1 | 800 | 212077_at | 0.4125 | 0.1585 | |
| GSE38129 | CALD1 | 800 | 212077_at | 0.0434 | 0.9186 | |
| GSE45670 | CALD1 | 800 | 212077_at | -1.3956 | 0.0002 | |
| GSE53622 | CALD1 | 800 | 14475 | -0.1506 | 0.3651 | |
| GSE53624 | CALD1 | 800 | 5689 | 0.2286 | 0.0223 | |
| GSE63941 | CALD1 | 800 | 212077_at | -4.1858 | 0.0026 | |
| GSE77861 | CALD1 | 800 | 212077_at | 1.6342 | 0.0072 | |
| GSE97050 | CALD1 | 800 | A_24_P921366 | -0.9711 | 0.1187 | |
| SRP007169 | CALD1 | 800 | RNAseq | 3.2669 | 0.0000 | |
| SRP008496 | CALD1 | 800 | RNAseq | 3.5058 | 0.0000 | |
| SRP064894 | CALD1 | 800 | RNAseq | 0.3508 | 0.2460 | |
| SRP133303 | CALD1 | 800 | RNAseq | 0.6288 | 0.1385 | |
| SRP159526 | CALD1 | 800 | RNAseq | -0.5740 | 0.1792 | |
| SRP193095 | CALD1 | 800 | RNAseq | 1.6276 | 0.0000 | |
| SRP219564 | CALD1 | 800 | RNAseq | -0.4939 | 0.5781 | |
| TCGA | CALD1 | 800 | RNAseq | -0.2472 | 0.0044 |
Upregulated datasets: 5; Downregulated datasets: 2.
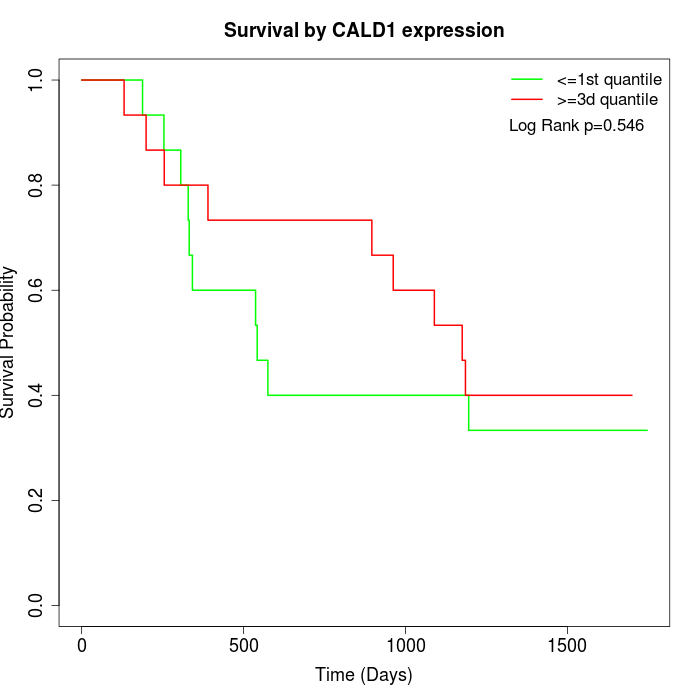
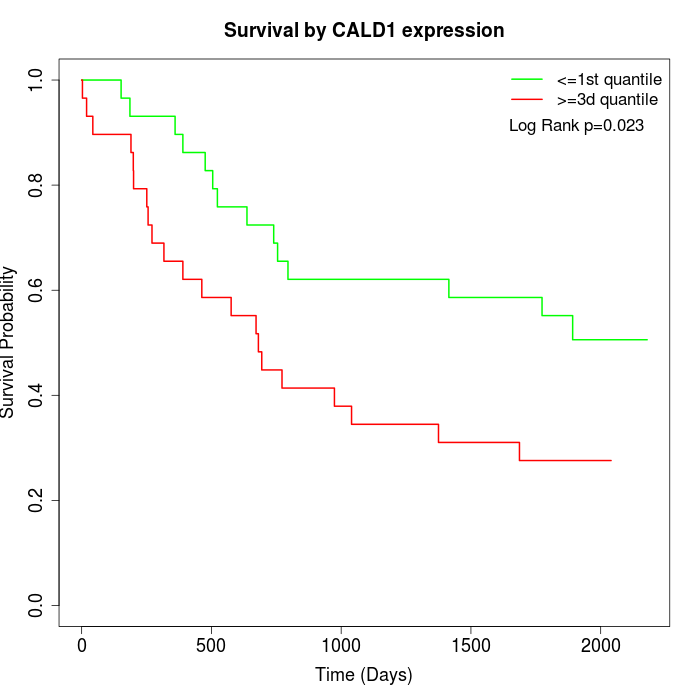
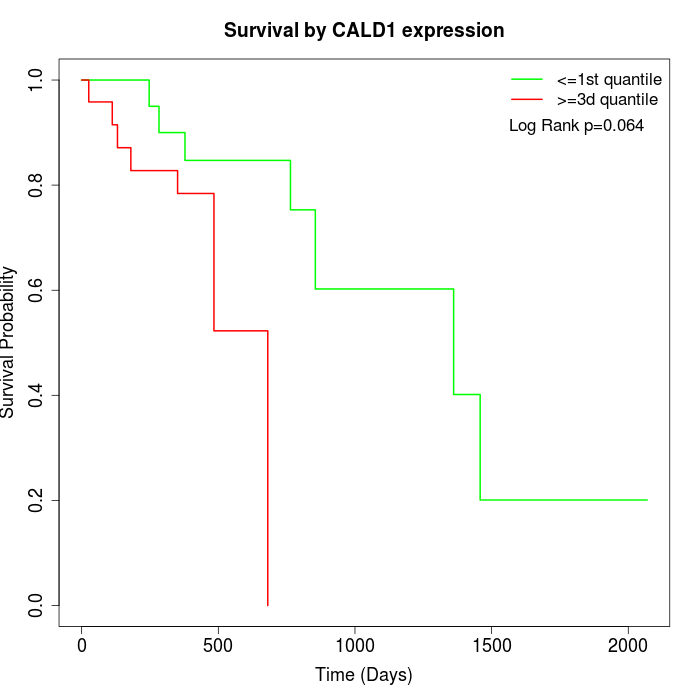
Survival by CALD1 expression:
Note: Click image to view full size file.
Copy number change of CALD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CALD1 | 800 | 6 | 2 | 22 | |
| GSE20123 | CALD1 | 800 | 6 | 2 | 22 | |
| GSE43470 | CALD1 | 800 | 2 | 3 | 38 | |
| GSE46452 | CALD1 | 800 | 7 | 2 | 50 | |
| GSE47630 | CALD1 | 800 | 7 | 4 | 29 | |
| GSE54993 | CALD1 | 800 | 3 | 5 | 62 | |
| GSE54994 | CALD1 | 800 | 6 | 8 | 39 | |
| GSE60625 | CALD1 | 800 | 0 | 0 | 11 | |
| GSE74703 | CALD1 | 800 | 2 | 3 | 31 | |
| GSE74704 | CALD1 | 800 | 2 | 2 | 16 | |
| TCGA | CALD1 | 800 | 29 | 23 | 44 |
Total number of gains: 70; Total number of losses: 54; Total Number of normals: 364.
Somatic mutations of CALD1:
Generating mutation plots.
Highly correlated genes for CALD1:
Showing top 20/1000 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CALD1 | TPM1 | 0.828666 | 13 | 0 | 12 |
| CALD1 | FERMT2 | 0.826364 | 13 | 0 | 13 |
| CALD1 | LAMA4 | 0.81873 | 12 | 0 | 12 |
| CALD1 | ZEB1 | 0.809301 | 3 | 0 | 3 |
| CALD1 | TPM2 | 0.800304 | 13 | 0 | 13 |
| CALD1 | FBN1 | 0.796666 | 11 | 0 | 11 |
| CALD1 | FILIP1L | 0.79582 | 13 | 0 | 13 |
| CALD1 | PMP22 | 0.791952 | 12 | 0 | 12 |
| CALD1 | TGFB1I1 | 0.783925 | 13 | 0 | 12 |
| CALD1 | ACTA2 | 0.779129 | 13 | 0 | 12 |
| CALD1 | ANTXR2 | 0.767304 | 9 | 0 | 9 |
| CALD1 | FN1 | 0.76588 | 11 | 0 | 10 |
| CALD1 | PDLIM3 | 0.765619 | 13 | 0 | 12 |
| CALD1 | SYNC | 0.763706 | 8 | 0 | 8 |
| CALD1 | DDR2 | 0.763169 | 12 | 0 | 12 |
| CALD1 | PALLD | 0.759289 | 13 | 0 | 11 |
| CALD1 | DCN | 0.756081 | 12 | 0 | 12 |
| CALD1 | MAP1B | 0.755718 | 11 | 0 | 10 |
| CALD1 | TMEM47 | 0.753978 | 13 | 0 | 11 |
| CALD1 | LINC01279 | 0.752835 | 4 | 0 | 4 |
For details and further investigation, click here