| Full name: SH2B adaptor protein 3 | Alias Symbol: LNK|IDDM20 | ||
| Type: protein-coding gene | Cytoband: 12q24.12 | ||
| Entrez ID: 10019 | HGNC ID: HGNC:29605 | Ensembl Gene: ENSG00000111252 | OMIM ID: 605093 |
| Drug and gene relationship at DGIdb | |||
SH2B3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway |
Expression of SH2B3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SH2B3 | 10019 | 203320_at | 0.3781 | 0.5291 | |
| GSE20347 | SH2B3 | 10019 | 203320_at | 0.3701 | 0.0010 | |
| GSE23400 | SH2B3 | 10019 | 203320_at | 0.2342 | 0.0075 | |
| GSE26886 | SH2B3 | 10019 | 203320_at | 0.5798 | 0.1282 | |
| GSE29001 | SH2B3 | 10019 | 203320_at | 0.3241 | 0.1353 | |
| GSE38129 | SH2B3 | 10019 | 203320_at | 0.2472 | 0.2740 | |
| GSE45670 | SH2B3 | 10019 | 203320_at | -0.2065 | 0.5562 | |
| GSE53622 | SH2B3 | 10019 | 10170 | 0.4550 | 0.0003 | |
| GSE53624 | SH2B3 | 10019 | 10170 | 0.6178 | 0.0000 | |
| GSE63941 | SH2B3 | 10019 | 203320_at | -2.2239 | 0.0010 | |
| GSE77861 | SH2B3 | 10019 | 203320_at | 0.2177 | 0.1963 | |
| GSE97050 | SH2B3 | 10019 | A_24_P201739 | 0.5265 | 0.1616 | |
| SRP007169 | SH2B3 | 10019 | RNAseq | 2.2839 | 0.0007 | |
| SRP008496 | SH2B3 | 10019 | RNAseq | 2.3618 | 0.0000 | |
| SRP064894 | SH2B3 | 10019 | RNAseq | 0.6553 | 0.0129 | |
| SRP133303 | SH2B3 | 10019 | RNAseq | 0.0369 | 0.8312 | |
| SRP159526 | SH2B3 | 10019 | RNAseq | 0.4578 | 0.1691 | |
| SRP193095 | SH2B3 | 10019 | RNAseq | 0.9834 | 0.0000 | |
| SRP219564 | SH2B3 | 10019 | RNAseq | 0.5889 | 0.2427 | |
| TCGA | SH2B3 | 10019 | RNAseq | 0.0681 | 0.4257 |
Upregulated datasets: 2; Downregulated datasets: 1.
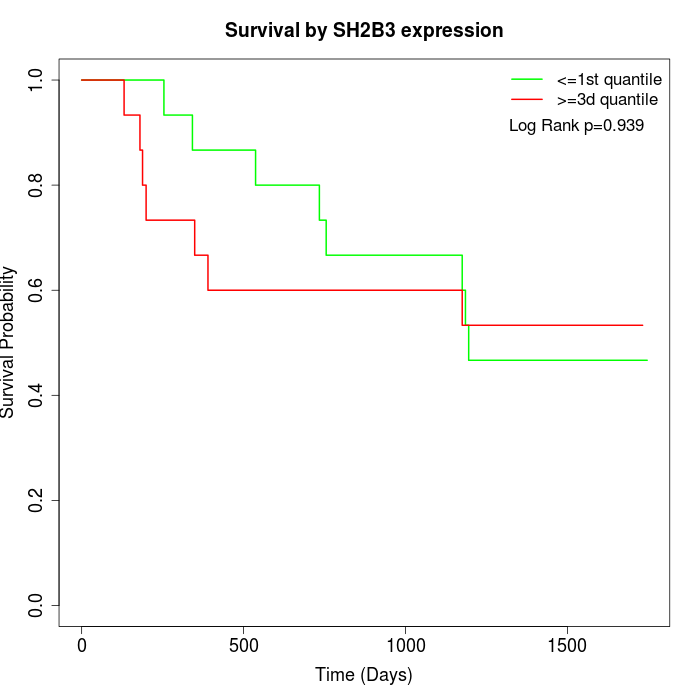
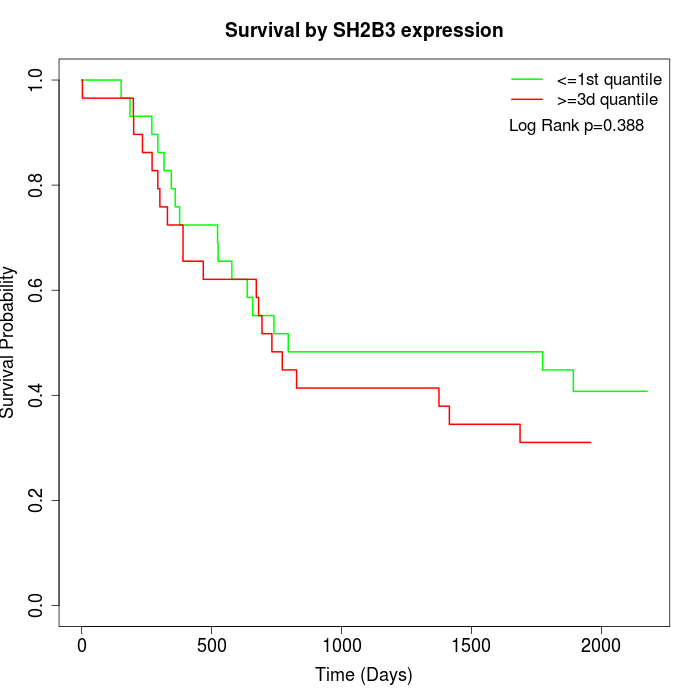
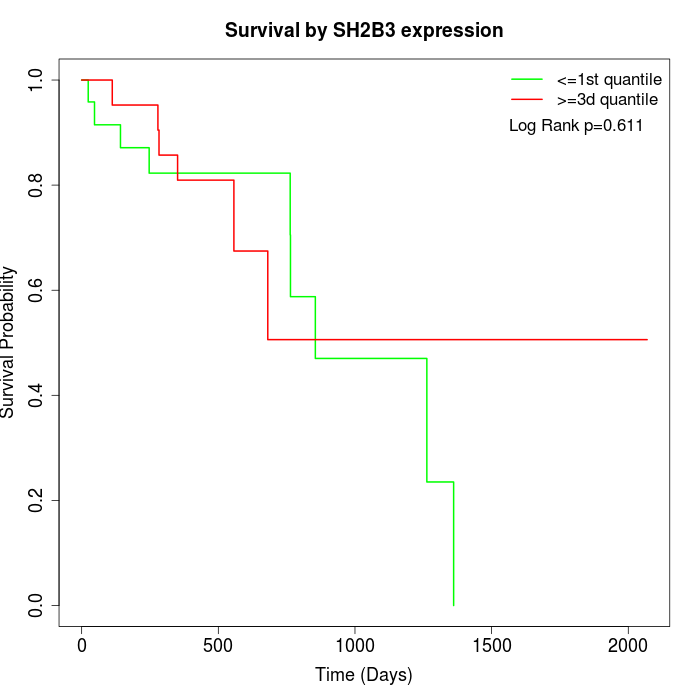
Survival by SH2B3 expression:
Note: Click image to view full size file.
Copy number change of SH2B3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SH2B3 | 10019 | 5 | 3 | 22 | |
| GSE20123 | SH2B3 | 10019 | 5 | 3 | 22 | |
| GSE43470 | SH2B3 | 10019 | 2 | 1 | 40 | |
| GSE46452 | SH2B3 | 10019 | 9 | 1 | 49 | |
| GSE47630 | SH2B3 | 10019 | 9 | 3 | 28 | |
| GSE54993 | SH2B3 | 10019 | 0 | 5 | 65 | |
| GSE54994 | SH2B3 | 10019 | 4 | 2 | 47 | |
| GSE60625 | SH2B3 | 10019 | 0 | 0 | 11 | |
| GSE74703 | SH2B3 | 10019 | 2 | 0 | 34 | |
| GSE74704 | SH2B3 | 10019 | 2 | 2 | 16 | |
| TCGA | SH2B3 | 10019 | 22 | 11 | 63 |
Total number of gains: 60; Total number of losses: 31; Total Number of normals: 397.
Somatic mutations of SH2B3:
Generating mutation plots.
Highly correlated genes for SH2B3:
Showing top 20/1355 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SH2B3 | FCGR3A | 0.807128 | 3 | 0 | 3 |
| SH2B3 | ENTPD4 | 0.786717 | 3 | 0 | 3 |
| SH2B3 | C19orf38 | 0.784284 | 3 | 0 | 3 |
| SH2B3 | CERKL | 0.761219 | 3 | 0 | 3 |
| SH2B3 | DERL3 | 0.759407 | 3 | 0 | 3 |
| SH2B3 | SAMM50 | 0.758727 | 3 | 0 | 3 |
| SH2B3 | GPR141 | 0.749682 | 3 | 0 | 3 |
| SH2B3 | ZNF18 | 0.746883 | 3 | 0 | 3 |
| SH2B3 | DEGS1 | 0.744301 | 3 | 0 | 3 |
| SH2B3 | TMEM200A | 0.742842 | 7 | 0 | 7 |
| SH2B3 | DRAM1 | 0.736844 | 13 | 0 | 12 |
| SH2B3 | ITGAM | 0.734832 | 9 | 0 | 8 |
| SH2B3 | SUSD1 | 0.728066 | 4 | 0 | 4 |
| SH2B3 | GLI2 | 0.724662 | 4 | 0 | 4 |
| SH2B3 | BICC1 | 0.720563 | 9 | 0 | 9 |
| SH2B3 | DNM3OS | 0.719149 | 3 | 0 | 3 |
| SH2B3 | KLHL21 | 0.717975 | 3 | 0 | 3 |
| SH2B3 | A1BG | 0.716341 | 3 | 0 | 3 |
| SH2B3 | TNFRSF1B | 0.715232 | 11 | 0 | 11 |
| SH2B3 | C1S | 0.712062 | 12 | 0 | 12 |
For details and further investigation, click here