| Full name: EGF like domain multiple 6 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: Xp22.2 | ||
| Entrez ID: 25975 | HGNC ID: HGNC:3235 | Ensembl Gene: ENSG00000198759 | OMIM ID: 300239 |
| Drug and gene relationship at DGIdb | |||
Expression of EGFL6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGFL6 | 25975 | 219454_at | 1.2470 | 0.3191 | |
| GSE20347 | EGFL6 | 25975 | 219454_at | 0.6393 | 0.0613 | |
| GSE23400 | EGFL6 | 25975 | 219454_at | 0.5958 | 0.0000 | |
| GSE26886 | EGFL6 | 25975 | 219454_at | -1.2463 | 0.0061 | |
| GSE29001 | EGFL6 | 25975 | 219454_at | -0.3394 | 0.5869 | |
| GSE38129 | EGFL6 | 25975 | 219454_at | 1.1534 | 0.0029 | |
| GSE45670 | EGFL6 | 25975 | 219454_at | 0.9649 | 0.0050 | |
| GSE53622 | EGFL6 | 25975 | 34526 | 1.2526 | 0.0000 | |
| GSE53624 | EGFL6 | 25975 | 34526 | 0.8676 | 0.0000 | |
| GSE63941 | EGFL6 | 25975 | 219454_at | 0.1652 | 0.9148 | |
| GSE77861 | EGFL6 | 25975 | 219454_at | -0.4839 | 0.4092 | |
| GSE97050 | EGFL6 | 25975 | A_33_P3290709 | 0.7979 | 0.1222 | |
| SRP007169 | EGFL6 | 25975 | RNAseq | 0.6403 | 0.2933 | |
| SRP008496 | EGFL6 | 25975 | RNAseq | 0.8420 | 0.0282 | |
| SRP064894 | EGFL6 | 25975 | RNAseq | 1.8784 | 0.0000 | |
| SRP133303 | EGFL6 | 25975 | RNAseq | 1.8825 | 0.0000 | |
| SRP159526 | EGFL6 | 25975 | RNAseq | -0.0404 | 0.9579 | |
| SRP219564 | EGFL6 | 25975 | RNAseq | 1.1176 | 0.1969 | |
| TCGA | EGFL6 | 25975 | RNAseq | 0.8153 | 0.0000 |
Upregulated datasets: 4; Downregulated datasets: 1.
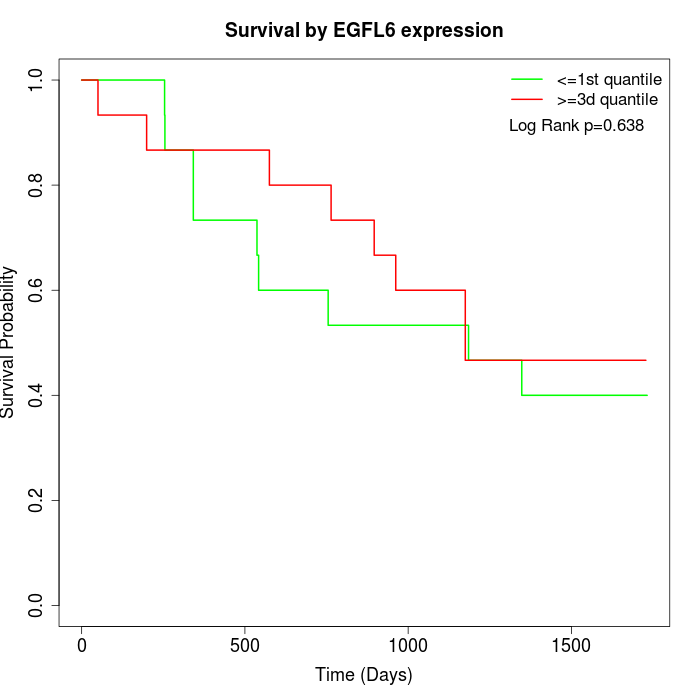
Survival by EGFL6 expression:
Note: Click image to view full size file.
Copy number change of EGFL6:
No record found for this gene.
Somatic mutations of EGFL6:
Generating mutation plots.
Highly correlated genes for EGFL6:
Showing top 20/362 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGFL6 | FAM117B | 0.70363 | 3 | 0 | 3 |
| EGFL6 | ALG10 | 0.702282 | 3 | 0 | 3 |
| EGFL6 | CNOT10 | 0.701966 | 3 | 0 | 3 |
| EGFL6 | PEX13 | 0.697932 | 3 | 0 | 3 |
| EGFL6 | SMURF1 | 0.689569 | 3 | 0 | 3 |
| EGFL6 | ISY1 | 0.676091 | 3 | 0 | 3 |
| EGFL6 | FERMT1 | 0.668829 | 4 | 0 | 4 |
| EGFL6 | GJB3 | 0.666124 | 3 | 0 | 3 |
| EGFL6 | C10orf55 | 0.664267 | 4 | 0 | 4 |
| EGFL6 | RCC2 | 0.6606 | 4 | 0 | 3 |
| EGFL6 | ZDHHC24 | 0.65554 | 3 | 0 | 3 |
| EGFL6 | CENPP | 0.652127 | 3 | 0 | 3 |
| EGFL6 | CYB5A | 0.647695 | 3 | 0 | 3 |
| EGFL6 | ARF3 | 0.646798 | 4 | 0 | 3 |
| EGFL6 | MSL3 | 0.645779 | 4 | 0 | 4 |
| EGFL6 | MRPL37 | 0.642123 | 5 | 0 | 5 |
| EGFL6 | PXMP2 | 0.639772 | 3 | 0 | 3 |
| EGFL6 | TRIM59 | 0.639025 | 4 | 0 | 4 |
| EGFL6 | RASSF9 | 0.637963 | 5 | 0 | 5 |
| EGFL6 | DPP3 | 0.631347 | 5 | 0 | 4 |
For details and further investigation, click here