| Full name: SMAD specific E3 ubiquitin protein ligase 1 | Alias Symbol: KIAA1625 | ||
| Type: protein-coding gene | Cytoband: 7q22.1 | ||
| Entrez ID: 57154 | HGNC ID: HGNC:16807 | Ensembl Gene: ENSG00000198742 | OMIM ID: 605568 |
| Drug and gene relationship at DGIdb | |||
SMURF1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04340 | Hedgehog signaling pathway | |
| hsa04350 | TGF-beta signaling pathway |
Expression of SMURF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMURF1 | 57154 | 212666_at | 0.0734 | 0.9231 | |
| GSE20347 | SMURF1 | 57154 | 212666_at | -0.1201 | 0.4701 | |
| GSE23400 | SMURF1 | 57154 | 212666_at | -0.0157 | 0.8917 | |
| GSE26886 | SMURF1 | 57154 | 212666_at | -0.5418 | 0.0169 | |
| GSE29001 | SMURF1 | 57154 | 212666_at | -0.4669 | 0.1205 | |
| GSE38129 | SMURF1 | 57154 | 212666_at | 0.0530 | 0.7887 | |
| GSE45670 | SMURF1 | 57154 | 212666_at | 0.0264 | 0.9283 | |
| GSE53622 | SMURF1 | 57154 | 48740 | -0.0991 | 0.4160 | |
| GSE53624 | SMURF1 | 57154 | 48740 | -0.1972 | 0.1493 | |
| GSE63941 | SMURF1 | 57154 | 212666_at | -0.5422 | 0.2577 | |
| GSE77861 | SMURF1 | 57154 | 212666_at | 0.0253 | 0.9202 | |
| GSE97050 | SMURF1 | 57154 | A_23_P353316 | -0.0827 | 0.8071 | |
| SRP007169 | SMURF1 | 57154 | RNAseq | -0.3534 | 0.3366 | |
| SRP008496 | SMURF1 | 57154 | RNAseq | -0.2400 | 0.3206 | |
| SRP064894 | SMURF1 | 57154 | RNAseq | -0.5045 | 0.0279 | |
| SRP133303 | SMURF1 | 57154 | RNAseq | 0.0674 | 0.7269 | |
| SRP159526 | SMURF1 | 57154 | RNAseq | -0.3442 | 0.3570 | |
| SRP193095 | SMURF1 | 57154 | RNAseq | 0.1039 | 0.4928 | |
| SRP219564 | SMURF1 | 57154 | RNAseq | -0.2830 | 0.5662 | |
| TCGA | SMURF1 | 57154 | RNAseq | 0.0035 | 0.9486 |
Upregulated datasets: 0; Downregulated datasets: 0.
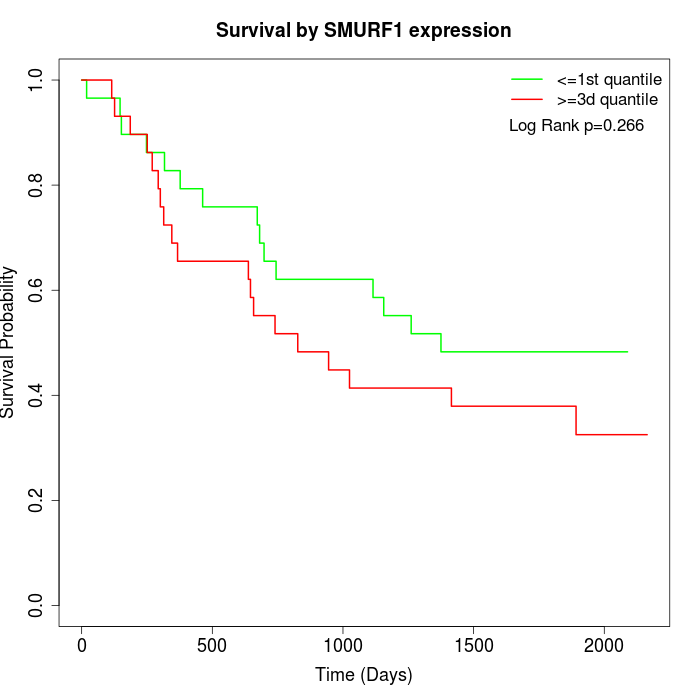
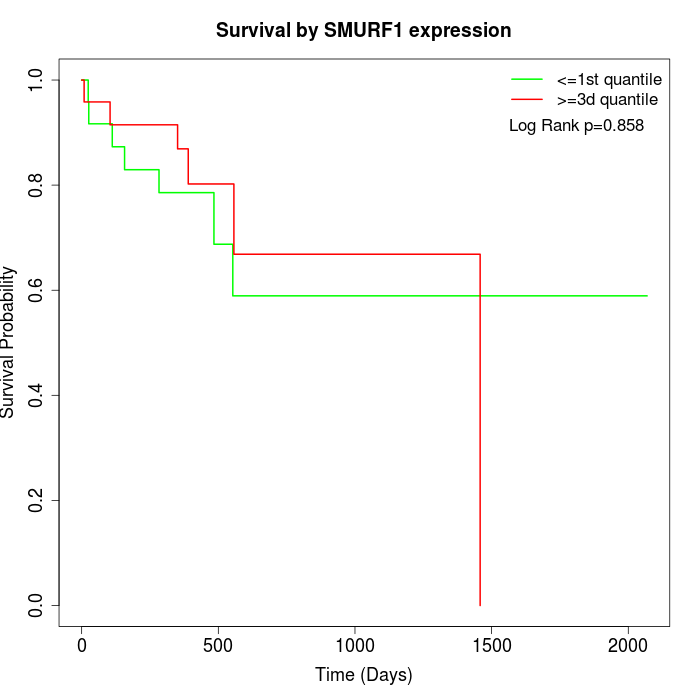
Survival by SMURF1 expression:
Note: Click image to view full size file.
Copy number change of SMURF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMURF1 | 57154 | 13 | 0 | 17 | |
| GSE20123 | SMURF1 | 57154 | 13 | 0 | 17 | |
| GSE43470 | SMURF1 | 57154 | 6 | 1 | 36 | |
| GSE46452 | SMURF1 | 57154 | 11 | 1 | 47 | |
| GSE47630 | SMURF1 | 57154 | 8 | 3 | 29 | |
| GSE54993 | SMURF1 | 57154 | 1 | 9 | 60 | |
| GSE54994 | SMURF1 | 57154 | 16 | 3 | 34 | |
| GSE60625 | SMURF1 | 57154 | 0 | 0 | 11 | |
| GSE74703 | SMURF1 | 57154 | 6 | 1 | 29 | |
| GSE74704 | SMURF1 | 57154 | 9 | 0 | 11 | |
| TCGA | SMURF1 | 57154 | 54 | 4 | 38 |
Total number of gains: 137; Total number of losses: 22; Total Number of normals: 329.
Somatic mutations of SMURF1:
Generating mutation plots.
Highly correlated genes for SMURF1:
Showing top 20/332 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMURF1 | DCTD | 0.800289 | 3 | 0 | 3 |
| SMURF1 | ACADVL | 0.794486 | 3 | 0 | 3 |
| SMURF1 | USP38 | 0.776697 | 3 | 0 | 3 |
| SMURF1 | PDIK1L | 0.772742 | 3 | 0 | 3 |
| SMURF1 | PCGF3 | 0.770867 | 3 | 0 | 3 |
| SMURF1 | PITHD1 | 0.769343 | 3 | 0 | 3 |
| SMURF1 | WDR77 | 0.758305 | 3 | 0 | 3 |
| SMURF1 | ASH2L | 0.757765 | 3 | 0 | 3 |
| SMURF1 | TMEM161B | 0.757636 | 3 | 0 | 3 |
| SMURF1 | ATP9A | 0.753053 | 3 | 0 | 3 |
| SMURF1 | PHLPP1 | 0.74798 | 3 | 0 | 3 |
| SMURF1 | COX15 | 0.745079 | 3 | 0 | 3 |
| SMURF1 | TMED2 | 0.743424 | 3 | 0 | 3 |
| SMURF1 | AP3M1 | 0.741602 | 3 | 0 | 3 |
| SMURF1 | USP40 | 0.739613 | 3 | 0 | 3 |
| SMURF1 | RETSAT | 0.737968 | 3 | 0 | 3 |
| SMURF1 | PINK1 | 0.734989 | 3 | 0 | 3 |
| SMURF1 | CPEB4 | 0.727394 | 3 | 0 | 3 |
| SMURF1 | CRADD | 0.721571 | 3 | 0 | 3 |
| SMURF1 | FBXW11 | 0.718555 | 3 | 0 | 3 |
For details and further investigation, click here