| Full name: epidermal growth factor receptor | Alias Symbol: ERBB1 | ||
| Type: protein-coding gene | Cytoband: 7p11.2 | ||
| Entrez ID: 1956 | HGNC ID: HGNC:3236 | Ensembl Gene: ENSG00000146648 | OMIM ID: 131550 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EGFR involved pathways:
Expression of EGFR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EGFR | 1956 | 201983_s_at | -0.0189 | 0.9813 | |
| GSE20347 | EGFR | 1956 | 201983_s_at | 0.6717 | 0.0241 | |
| GSE23400 | EGFR | 1956 | 201983_s_at | 0.7661 | 0.0001 | |
| GSE26886 | EGFR | 1956 | 201983_s_at | 1.3177 | 0.0001 | |
| GSE29001 | EGFR | 1956 | 201983_s_at | 0.9551 | 0.0866 | |
| GSE38129 | EGFR | 1956 | 201983_s_at | 0.5322 | 0.1130 | |
| GSE45670 | EGFR | 1956 | 224999_at | 0.0918 | 0.7140 | |
| GSE53622 | EGFR | 1956 | 47004 | 0.7220 | 0.0000 | |
| GSE53624 | EGFR | 1956 | 47004 | 0.7309 | 0.0000 | |
| GSE63941 | EGFR | 1956 | 201983_s_at | 2.9877 | 0.0010 | |
| GSE77861 | EGFR | 1956 | 201983_s_at | 0.9697 | 0.0637 | |
| GSE97050 | EGFR | 1956 | A_23_P215790 | 0.4537 | 0.2308 | |
| SRP008496 | EGFR | 1956 | RNAseq | 3.4615 | 0.0001 | |
| SRP064894 | EGFR | 1956 | RNAseq | 1.0714 | 0.0053 | |
| SRP133303 | EGFR | 1956 | RNAseq | 0.3125 | 0.2705 | |
| SRP159526 | EGFR | 1956 | RNAseq | 0.6850 | 0.0597 | |
| SRP193095 | EGFR | 1956 | RNAseq | 1.0429 | 0.0002 | |
| SRP219564 | EGFR | 1956 | RNAseq | 0.3769 | 0.4021 | |
| TCGA | EGFR | 1956 | RNAseq | 0.2376 | 0.0220 |
Upregulated datasets: 5; Downregulated datasets: 0.
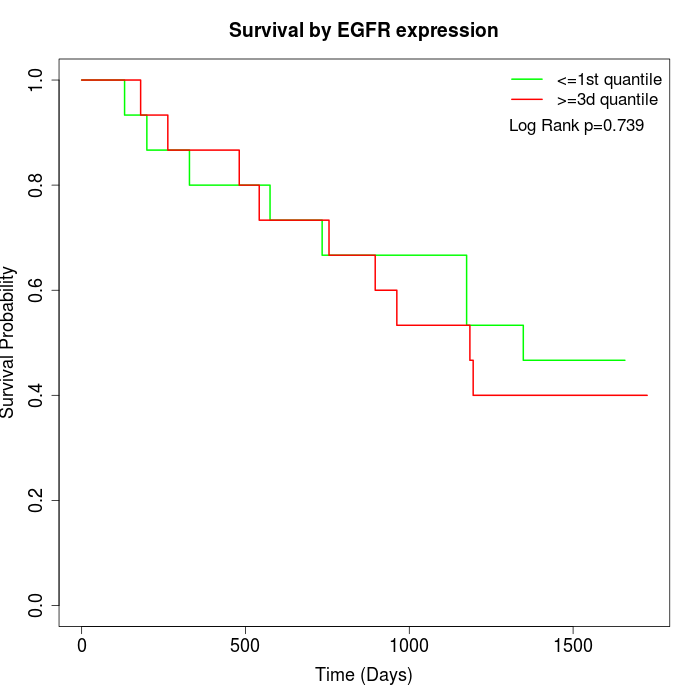
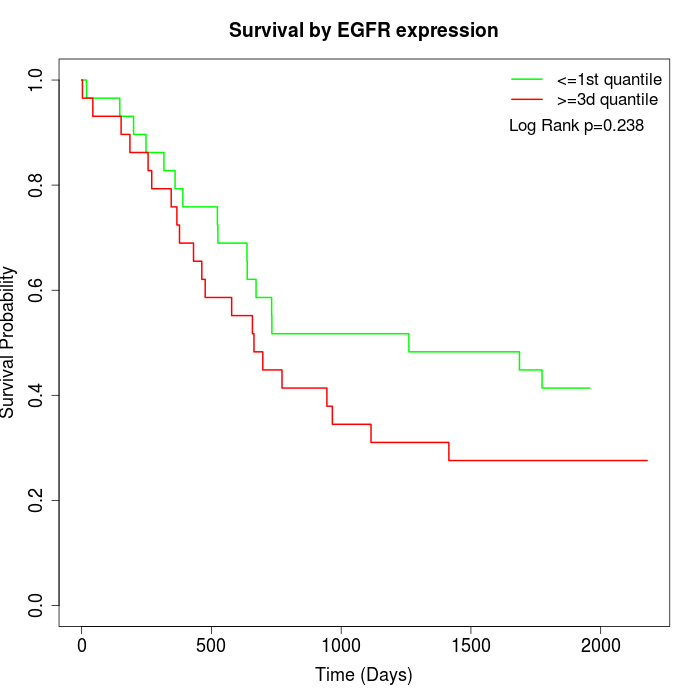
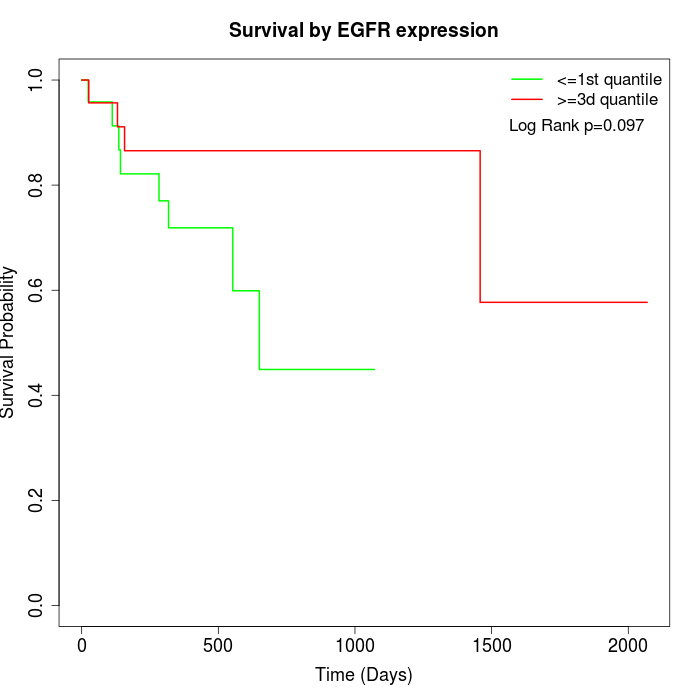
Survival by EGFR expression:
Note: Click image to view full size file.
Copy number change of EGFR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EGFR | 1956 | 14 | 0 | 16 | |
| GSE20123 | EGFR | 1956 | 12 | 0 | 18 | |
| GSE43470 | EGFR | 1956 | 6 | 0 | 37 | |
| GSE46452 | EGFR | 1956 | 13 | 0 | 46 | |
| GSE47630 | EGFR | 1956 | 10 | 1 | 29 | |
| GSE54993 | EGFR | 1956 | 0 | 11 | 59 | |
| GSE54994 | EGFR | 1956 | 18 | 1 | 34 | |
| GSE60625 | EGFR | 1956 | 0 | 0 | 11 | |
| GSE74703 | EGFR | 1956 | 6 | 0 | 30 | |
| GSE74704 | EGFR | 1956 | 6 | 0 | 14 | |
| TCGA | EGFR | 1956 | 57 | 5 | 34 |
Total number of gains: 142; Total number of losses: 18; Total Number of normals: 328.
Somatic mutations of EGFR:
Generating mutation plots.
Highly correlated genes for EGFR:
Showing top 20/583 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EGFR | CHST3 | 0.725451 | 3 | 0 | 3 |
| EGFR | LRRC8A | 0.698022 | 3 | 0 | 3 |
| EGFR | MB21D2 | 0.697188 | 3 | 0 | 3 |
| EGFR | RILPL1 | 0.695636 | 3 | 0 | 3 |
| EGFR | RRNAD1 | 0.691063 | 3 | 0 | 3 |
| EGFR | ELOVL7 | 0.685698 | 4 | 0 | 4 |
| EGFR | SLC41A1 | 0.682154 | 3 | 0 | 3 |
| EGFR | ALKBH6 | 0.681871 | 3 | 0 | 3 |
| EGFR | TMEM160 | 0.679864 | 3 | 0 | 3 |
| EGFR | TMEM138 | 0.677279 | 4 | 0 | 4 |
| EGFR | CHCHD6 | 0.676066 | 3 | 0 | 3 |
| EGFR | DHX36 | 0.674033 | 5 | 0 | 4 |
| EGFR | C12orf45 | 0.669599 | 3 | 0 | 3 |
| EGFR | MRPS26 | 0.66652 | 3 | 0 | 3 |
| EGFR | CYTH3 | 0.665647 | 4 | 0 | 3 |
| EGFR | PHF14 | 0.664786 | 5 | 0 | 5 |
| EGFR | CNTN1 | 0.660293 | 3 | 0 | 3 |
| EGFR | HSPA13 | 0.656189 | 3 | 0 | 3 |
| EGFR | CMTM4 | 0.655709 | 3 | 0 | 3 |
| EGFR | TRMT2B | 0.654338 | 4 | 0 | 4 |
For details and further investigation, click here