| Full name: Fas associated factor family member 2 | Alias Symbol: ETEA|KIAA0887|UBXN3B | ||
| Type: protein-coding gene | Cytoband: 5q35.2 | ||
| Entrez ID: 23197 | HGNC ID: HGNC:24666 | Ensembl Gene: ENSG00000113194 | OMIM ID: 616935 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of FAF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAF2 | 23197 | 212108_at | 0.3884 | 0.3877 | |
| GSE20347 | FAF2 | 23197 | 212108_at | 0.0449 | 0.6558 | |
| GSE23400 | FAF2 | 23197 | 212108_at | 0.1733 | 0.0053 | |
| GSE26886 | FAF2 | 23197 | 212108_at | 0.6436 | 0.0002 | |
| GSE29001 | FAF2 | 23197 | 212108_at | 0.1979 | 0.6237 | |
| GSE38129 | FAF2 | 23197 | 212108_at | 0.1026 | 0.2967 | |
| GSE45670 | FAF2 | 23197 | 212108_at | 0.1212 | 0.5089 | |
| GSE53622 | FAF2 | 23197 | 90736 | 0.2244 | 0.0011 | |
| GSE53624 | FAF2 | 23197 | 90736 | 0.2276 | 0.0000 | |
| GSE63941 | FAF2 | 23197 | 212108_at | 0.1127 | 0.8190 | |
| GSE77861 | FAF2 | 23197 | 212108_at | 0.3200 | 0.0744 | |
| GSE97050 | FAF2 | 23197 | A_24_P294719 | 0.1170 | 0.5911 | |
| SRP007169 | FAF2 | 23197 | RNAseq | 0.2083 | 0.5662 | |
| SRP008496 | FAF2 | 23197 | RNAseq | -0.0494 | 0.8312 | |
| SRP064894 | FAF2 | 23197 | RNAseq | 0.2087 | 0.1276 | |
| SRP133303 | FAF2 | 23197 | RNAseq | 0.5049 | 0.0093 | |
| SRP159526 | FAF2 | 23197 | RNAseq | 0.0658 | 0.8250 | |
| SRP193095 | FAF2 | 23197 | RNAseq | 0.2492 | 0.0199 | |
| SRP219564 | FAF2 | 23197 | RNAseq | 0.3861 | 0.3052 | |
| TCGA | FAF2 | 23197 | RNAseq | 0.0453 | 0.3158 |
Upregulated datasets: 0; Downregulated datasets: 0.
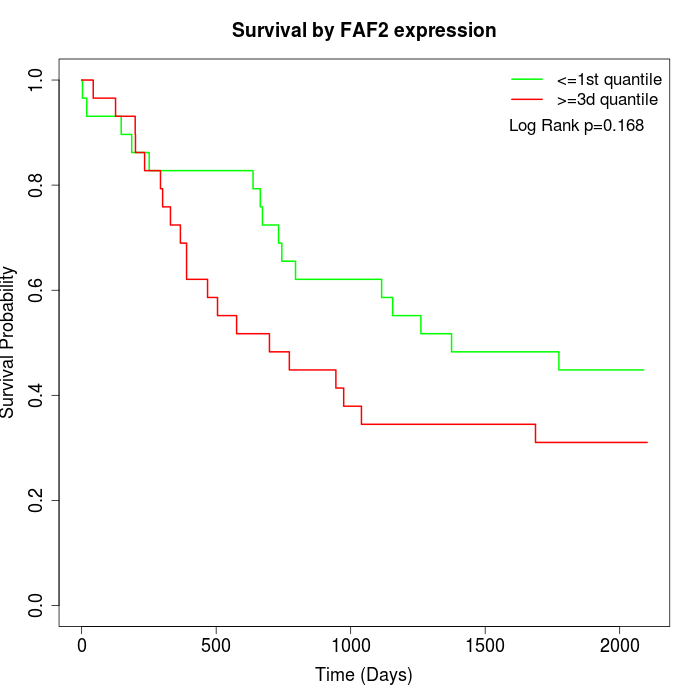
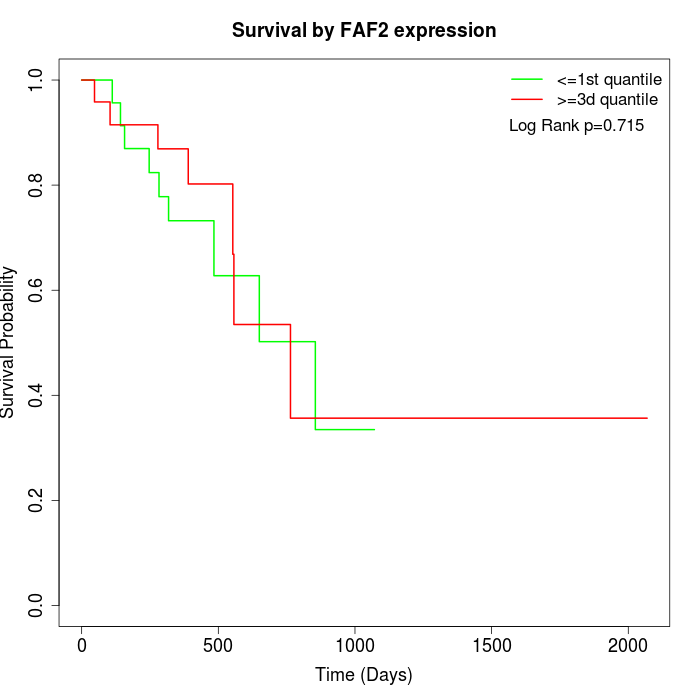
Survival by FAF2 expression:
Note: Click image to view full size file.
Copy number change of FAF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAF2 | 23197 | 2 | 13 | 15 | |
| GSE20123 | FAF2 | 23197 | 2 | 13 | 15 | |
| GSE43470 | FAF2 | 23197 | 1 | 11 | 31 | |
| GSE46452 | FAF2 | 23197 | 0 | 27 | 32 | |
| GSE47630 | FAF2 | 23197 | 0 | 20 | 20 | |
| GSE54993 | FAF2 | 23197 | 10 | 2 | 58 | |
| GSE54994 | FAF2 | 23197 | 2 | 17 | 34 | |
| GSE60625 | FAF2 | 23197 | 1 | 0 | 10 | |
| GSE74703 | FAF2 | 23197 | 1 | 7 | 28 | |
| GSE74704 | FAF2 | 23197 | 1 | 6 | 13 | |
| TCGA | FAF2 | 23197 | 7 | 35 | 54 |
Total number of gains: 27; Total number of losses: 151; Total Number of normals: 310.
Somatic mutations of FAF2:
Generating mutation plots.
Highly correlated genes for FAF2:
Showing top 20/798 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAF2 | SCRN3 | 0.854742 | 3 | 0 | 3 |
| FAF2 | NFE2L1 | 0.835972 | 3 | 0 | 3 |
| FAF2 | B3GNT9 | 0.801184 | 3 | 0 | 3 |
| FAF2 | FAM168A | 0.794538 | 3 | 0 | 3 |
| FAF2 | XIAP | 0.787588 | 3 | 0 | 3 |
| FAF2 | MASTL | 0.783277 | 3 | 0 | 3 |
| FAF2 | HNRNPUL1 | 0.781528 | 3 | 0 | 3 |
| FAF2 | EP300 | 0.774513 | 3 | 0 | 3 |
| FAF2 | KIAA1191 | 0.774488 | 3 | 0 | 3 |
| FAF2 | THOC2 | 0.77279 | 3 | 0 | 3 |
| FAF2 | PPP6R3 | 0.75694 | 4 | 0 | 4 |
| FAF2 | AP3D1 | 0.754739 | 4 | 0 | 4 |
| FAF2 | LACTB | 0.751211 | 4 | 0 | 4 |
| FAF2 | GLT8D1 | 0.750979 | 3 | 0 | 3 |
| FAF2 | SPIRE1 | 0.750745 | 4 | 0 | 3 |
| FAF2 | CNOT2 | 0.745737 | 4 | 0 | 4 |
| FAF2 | ROMO1 | 0.744927 | 3 | 0 | 3 |
| FAF2 | ZBED1 | 0.744003 | 3 | 0 | 3 |
| FAF2 | IPP | 0.743398 | 3 | 0 | 3 |
| FAF2 | CTDSPL2 | 0.742199 | 3 | 0 | 3 |
For details and further investigation, click here