| Full name: family with sequence similarity 166 member A | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 9q34.3 | ||
| Entrez ID: 401565 | HGNC ID: HGNC:33818 | Ensembl Gene: ENSG00000188163 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of FAM166A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAM166A | 401565 | 1562778_at | -0.0172 | 0.9599 | |
| GSE26886 | FAM166A | 401565 | 1562778_at | 0.0153 | 0.9358 | |
| GSE45670 | FAM166A | 401565 | 1562778_at | 0.0025 | 0.9856 | |
| GSE63941 | FAM166A | 401565 | 1562778_at | 0.1056 | 0.4137 | |
| GSE77861 | FAM166A | 401565 | 1562778_at | -0.1578 | 0.2943 | |
| GSE97050 | FAM166A | 401565 | A_33_P3295870 | -0.3254 | 0.2827 | |
| TCGA | FAM166A | 401565 | RNAseq | 0.3739 | 0.4992 |
Upregulated datasets: 0; Downregulated datasets: 0.
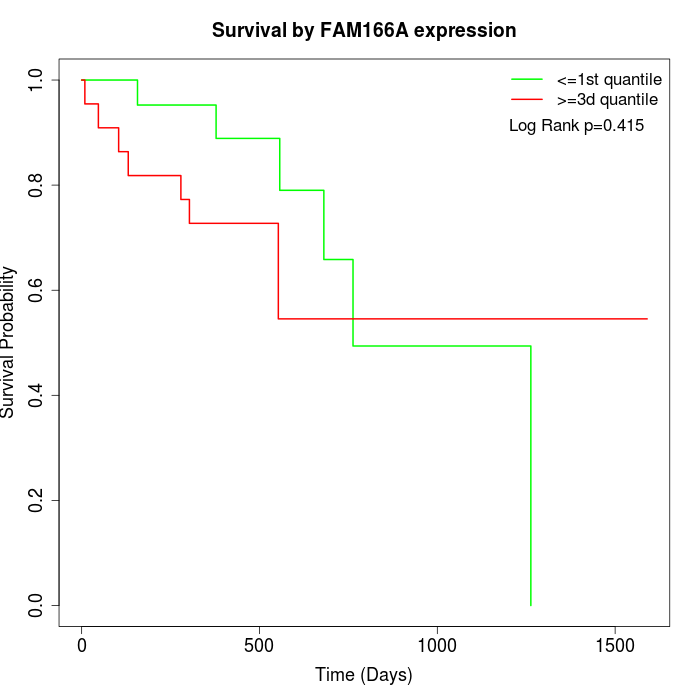
Survival by FAM166A expression:
Note: Click image to view full size file.
Copy number change of FAM166A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAM166A | 401565 | 5 | 7 | 18 | |
| GSE20123 | FAM166A | 401565 | 5 | 7 | 18 | |
| GSE43470 | FAM166A | 401565 | 3 | 7 | 33 | |
| GSE46452 | FAM166A | 401565 | 6 | 13 | 40 | |
| GSE47630 | FAM166A | 401565 | 6 | 15 | 19 | |
| GSE54993 | FAM166A | 401565 | 3 | 3 | 64 | |
| GSE54994 | FAM166A | 401565 | 12 | 7 | 34 | |
| GSE60625 | FAM166A | 401565 | 0 | 0 | 11 | |
| GSE74703 | FAM166A | 401565 | 3 | 5 | 28 | |
| GSE74704 | FAM166A | 401565 | 3 | 5 | 12 | |
| TCGA | FAM166A | 401565 | 30 | 23 | 43 |
Total number of gains: 76; Total number of losses: 92; Total Number of normals: 320.
Somatic mutations of FAM166A:
Generating mutation plots.
Highly correlated genes for FAM166A:
Showing top 20/109 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAM166A | SEMA4A | 0.775977 | 3 | 0 | 3 |
| FAM166A | TRPC4 | 0.760971 | 3 | 0 | 3 |
| FAM166A | RSPH3 | 0.756557 | 3 | 0 | 3 |
| FAM166A | C2CD2L | 0.730083 | 3 | 0 | 3 |
| FAM166A | PPP1R1B | 0.726873 | 3 | 0 | 3 |
| FAM166A | POU3F4 | 0.724407 | 3 | 0 | 3 |
| FAM166A | TSC22D4 | 0.71978 | 3 | 0 | 3 |
| FAM166A | ZNF681 | 0.71195 | 3 | 0 | 3 |
| FAM166A | ANO8 | 0.703401 | 3 | 0 | 3 |
| FAM166A | SNCG | 0.689103 | 3 | 0 | 3 |
| FAM166A | TMEM38A | 0.681783 | 3 | 0 | 3 |
| FAM166A | FBXO10 | 0.68038 | 3 | 0 | 3 |
| FAM166A | HECW2 | 0.680322 | 3 | 0 | 3 |
| FAM166A | ELMOD3 | 0.677643 | 3 | 0 | 3 |
| FAM166A | TMEM214 | 0.676839 | 3 | 0 | 3 |
| FAM166A | RFFL | 0.67619 | 3 | 0 | 3 |
| FAM166A | AMOTL2 | 0.669919 | 3 | 0 | 3 |
| FAM166A | COL4A4 | 0.667633 | 3 | 0 | 3 |
| FAM166A | ZNF345 | 0.667363 | 3 | 0 | 3 |
| FAM166A | CD200 | 0.667156 | 3 | 0 | 3 |
For details and further investigation, click here