| Full name: family with sequence similarity 66 member D | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 8p23.1 | ||
| Entrez ID: 100132923 | HGNC ID: HGNC:24159 | Ensembl Gene: ENSG00000255052 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of FAM66D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAM66D | 100132923 | 222194_at | 0.0326 | 0.9200 | |
| GSE20347 | FAM66D | 100132923 | 222194_at | 0.1288 | 0.1217 | |
| GSE23400 | FAM66D | 100132923 | 222194_at | -0.0202 | 0.4702 | |
| GSE26886 | FAM66D | 100132923 | 222194_at | 0.0083 | 0.9535 | |
| GSE29001 | FAM66D | 100132923 | 222194_at | 0.0326 | 0.8438 | |
| GSE38129 | FAM66D | 100132923 | 222194_at | 0.0366 | 0.6109 | |
| GSE45670 | FAM66D | 100132923 | 222194_at | 0.0351 | 0.6524 | |
| GSE63941 | FAM66D | 100132923 | 222194_at | 0.1028 | 0.5259 | |
| GSE77861 | FAM66D | 100132923 | 222194_at | -0.0279 | 0.7165 | |
| GSE97050 | FAM66D | A_21_P0013535 | 0.0181 | 0.9500 | ||
| TCGA | FAM66D | 100132923 | RNAseq | -0.1498 | 0.7671 |
Upregulated datasets: 0; Downregulated datasets: 0.
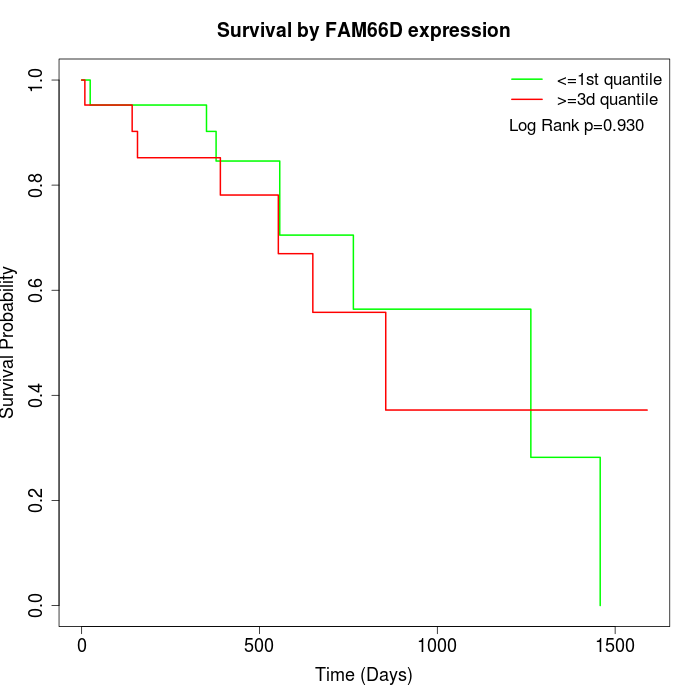
Survival by FAM66D expression:
Note: Click image to view full size file.
Copy number change of FAM66D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAM66D | 100132923 | 4 | 10 | 16 | |
| GSE20123 | FAM66D | 100132923 | 4 | 10 | 16 | |
| GSE43470 | FAM66D | 100132923 | 4 | 9 | 30 | |
| GSE46452 | FAM66D | 100132923 | 12 | 14 | 33 | |
| GSE47630 | FAM66D | 100132923 | 10 | 7 | 23 | |
| GSE54993 | FAM66D | 100132923 | 3 | 14 | 53 | |
| GSE54994 | FAM66D | 100132923 | 9 | 16 | 28 | |
| GSE60625 | FAM66D | 100132923 | 3 | 0 | 8 | |
| GSE74703 | FAM66D | 100132923 | 4 | 7 | 25 | |
| GSE74704 | FAM66D | 100132923 | 3 | 7 | 10 | |
| TCGA | FAM66D | 100132923 | 14 | 39 | 43 |
Total number of gains: 70; Total number of losses: 133; Total Number of normals: 285.
Somatic mutations of FAM66D:
Generating mutation plots.
Highly correlated genes for FAM66D:
Showing top 20/441 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAM66D | DAB1 | 0.713022 | 4 | 0 | 4 |
| FAM66D | KRT84 | 0.708091 | 3 | 0 | 3 |
| FAM66D | SLC30A3 | 0.70799 | 5 | 0 | 5 |
| FAM66D | SGK2 | 0.707208 | 6 | 0 | 6 |
| FAM66D | MUC2 | 0.698333 | 3 | 0 | 3 |
| FAM66D | TAL1 | 0.682177 | 5 | 0 | 5 |
| FAM66D | LCT | 0.671037 | 5 | 0 | 4 |
| FAM66D | RPGRIP1 | 0.667035 | 4 | 0 | 3 |
| FAM66D | RUNX2 | 0.663593 | 5 | 0 | 4 |
| FAM66D | HECW1 | 0.663366 | 4 | 0 | 3 |
| FAM66D | PHOX2B | 0.66322 | 5 | 0 | 5 |
| FAM66D | CNTN2 | 0.658386 | 5 | 0 | 5 |
| FAM66D | WRNIP1 | 0.657399 | 4 | 0 | 4 |
| FAM66D | TNFSF8 | 0.653017 | 4 | 0 | 3 |
| FAM66D | PPP1R37 | 0.651316 | 4 | 0 | 4 |
| FAM66D | CALCB | 0.650027 | 3 | 0 | 3 |
| FAM66D | NTNG1 | 0.647194 | 4 | 0 | 4 |
| FAM66D | RFPL2 | 0.64505 | 5 | 0 | 4 |
| FAM66D | DENND6B | 0.644372 | 4 | 0 | 4 |
| FAM66D | CLDN16 | 0.64355 | 5 | 0 | 4 |
For details and further investigation, click here