| Full name: Fas cell surface death receptor | Alias Symbol: CD95|APO-1 | ||
| Type: protein-coding gene | Cytoband: 10q23.31 | ||
| Entrez ID: 355 | HGNC ID: HGNC:11920 | Ensembl Gene: ENSG00000026103 | OMIM ID: 134637 |
| Drug and gene relationship at DGIdb | |||
FAS involved pathways:
Expression of FAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FAS | 355 | 204780_s_at | -0.4257 | 0.2480 | |
| GSE20347 | FAS | 355 | 204780_s_at | -1.2644 | 0.0001 | |
| GSE23400 | FAS | 355 | 204780_s_at | -0.0002 | 0.9986 | |
| GSE26886 | FAS | 355 | 204780_s_at | -0.7068 | 0.0908 | |
| GSE29001 | FAS | 355 | 204780_s_at | -0.6933 | 0.2171 | |
| GSE38129 | FAS | 355 | 204780_s_at | -0.6921 | 0.0049 | |
| GSE45670 | FAS | 355 | 204780_s_at | -0.6419 | 0.0076 | |
| GSE53622 | FAS | 355 | 49482 | -0.3860 | 0.0005 | |
| GSE53624 | FAS | 355 | 49482 | -0.7724 | 0.0000 | |
| GSE63941 | FAS | 355 | 204780_s_at | -2.5620 | 0.0316 | |
| GSE77861 | FAS | 355 | 204781_s_at | -0.4827 | 0.1677 | |
| GSE97050 | FAS | 355 | A_23_P63896 | 0.1655 | 0.7358 | |
| SRP007169 | FAS | 355 | RNAseq | -1.6600 | 0.0016 | |
| SRP008496 | FAS | 355 | RNAseq | -1.1453 | 0.0001 | |
| SRP064894 | FAS | 355 | RNAseq | -0.2306 | 0.3753 | |
| SRP133303 | FAS | 355 | RNAseq | -0.4959 | 0.0474 | |
| SRP159526 | FAS | 355 | RNAseq | -0.9029 | 0.0245 | |
| SRP193095 | FAS | 355 | RNAseq | -0.7175 | 0.0000 | |
| SRP219564 | FAS | 355 | RNAseq | 0.2469 | 0.3778 | |
| TCGA | FAS | 355 | RNAseq | 0.2685 | 0.0072 |
Upregulated datasets: 0; Downregulated datasets: 4.
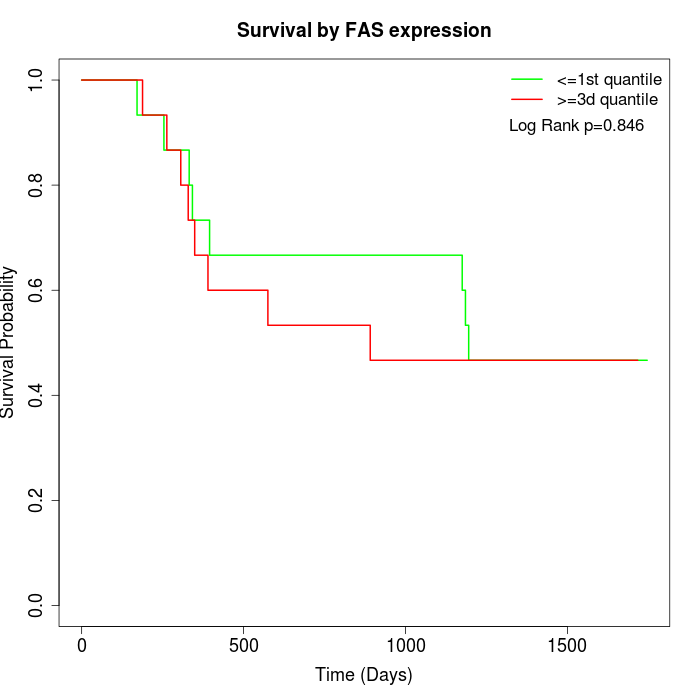
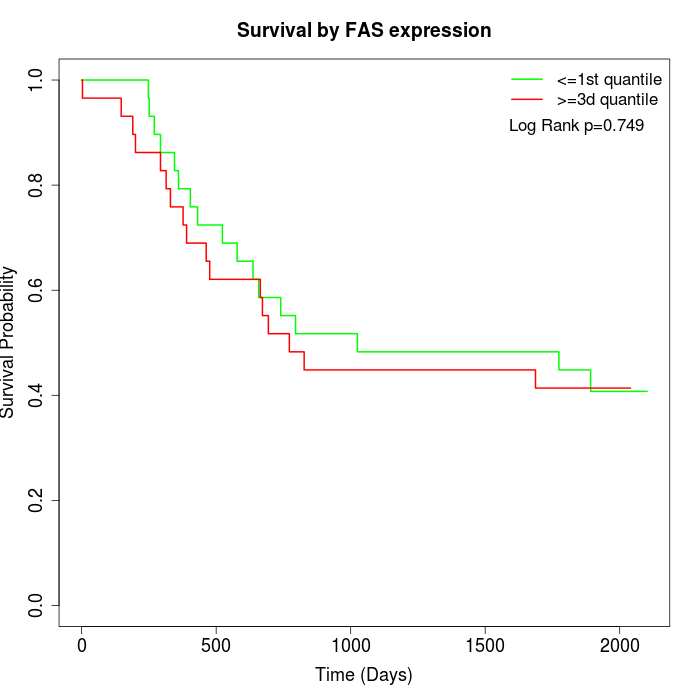
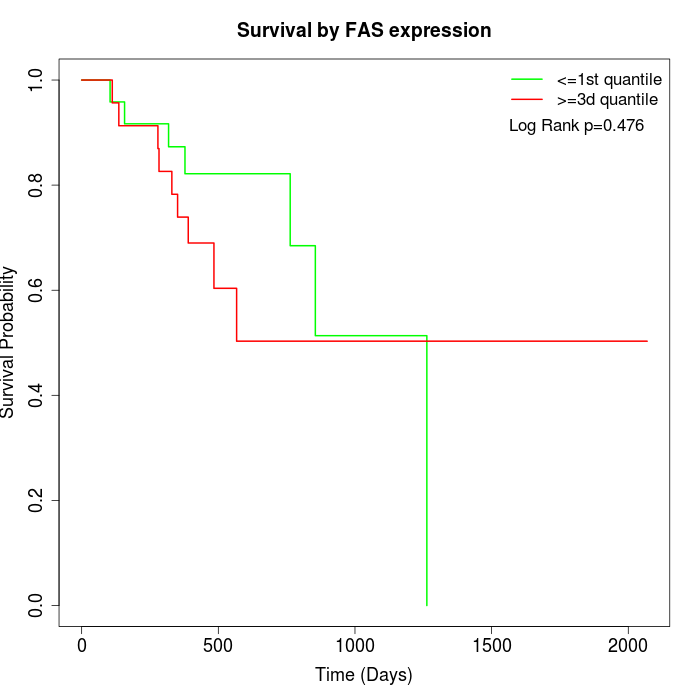
Survival by FAS expression:
Note: Click image to view full size file.
Copy number change of FAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FAS | 355 | 2 | 7 | 21 | |
| GSE20123 | FAS | 355 | 2 | 6 | 22 | |
| GSE43470 | FAS | 355 | 0 | 8 | 35 | |
| GSE46452 | FAS | 355 | 0 | 11 | 48 | |
| GSE47630 | FAS | 355 | 2 | 14 | 24 | |
| GSE54993 | FAS | 355 | 7 | 0 | 63 | |
| GSE54994 | FAS | 355 | 1 | 11 | 41 | |
| GSE60625 | FAS | 355 | 0 | 0 | 11 | |
| GSE74703 | FAS | 355 | 0 | 6 | 30 | |
| GSE74704 | FAS | 355 | 1 | 4 | 15 | |
| TCGA | FAS | 355 | 7 | 27 | 62 |
Total number of gains: 22; Total number of losses: 94; Total Number of normals: 372.
Somatic mutations of FAS:
Generating mutation plots.
Highly correlated genes for FAS:
Showing top 20/577 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FAS | CCNC | 0.737852 | 3 | 0 | 3 |
| FAS | LACTB | 0.734049 | 3 | 0 | 3 |
| FAS | SIKE1 | 0.729875 | 3 | 0 | 3 |
| FAS | ZNF267 | 0.715042 | 3 | 0 | 3 |
| FAS | PTGR1 | 0.712579 | 3 | 0 | 3 |
| FAS | ARHGAP9 | 0.69594 | 3 | 0 | 3 |
| FAS | ECSCR | 0.690654 | 3 | 0 | 3 |
| FAS | ZNF436 | 0.679262 | 4 | 0 | 3 |
| FAS | HNRNPUL2 | 0.677068 | 3 | 0 | 3 |
| FAS | SAMD3 | 0.675031 | 3 | 0 | 3 |
| FAS | AQR | 0.673929 | 3 | 0 | 3 |
| FAS | TPSD1 | 0.671426 | 3 | 0 | 3 |
| FAS | ST8SIA4 | 0.662113 | 3 | 0 | 3 |
| FAS | SCARF1 | 0.660656 | 4 | 0 | 3 |
| FAS | TAGAP | 0.660026 | 4 | 0 | 3 |
| FAS | PRMT6 | 0.659156 | 3 | 0 | 3 |
| FAS | QSER1 | 0.656414 | 3 | 0 | 3 |
| FAS | GPR183 | 0.655904 | 4 | 0 | 4 |
| FAS | CCL11 | 0.654002 | 4 | 0 | 3 |
| FAS | PEX11B | 0.6532 | 5 | 0 | 5 |
For details and further investigation, click here