| Full name: cyclin C | Alias Symbol: CycC | ||
| Type: protein-coding gene | Cytoband: 6q16.2 | ||
| Entrez ID: 892 | HGNC ID: HGNC:1581 | Ensembl Gene: ENSG00000112237 | OMIM ID: 123838 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CCNC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CCNC | 892 | 201955_at | -0.3740 | 0.4961 | |
| GSE20347 | CCNC | 892 | 201955_at | -0.2951 | 0.0319 | |
| GSE23400 | CCNC | 892 | 201955_at | -0.1395 | 0.1325 | |
| GSE26886 | CCNC | 892 | 201955_at | -0.2167 | 0.3625 | |
| GSE29001 | CCNC | 892 | 201955_at | -0.3045 | 0.4528 | |
| GSE38129 | CCNC | 892 | 201955_at | -0.0916 | 0.5481 | |
| GSE45670 | CCNC | 892 | 201955_at | -0.1490 | 0.1532 | |
| GSE53622 | CCNC | 892 | 47973 | -0.3147 | 0.0001 | |
| GSE53624 | CCNC | 892 | 47973 | -0.2914 | 0.0046 | |
| GSE63941 | CCNC | 892 | 201955_at | 0.7135 | 0.0859 | |
| GSE77861 | CCNC | 892 | 201955_at | -0.3807 | 0.3072 | |
| GSE97050 | CCNC | 892 | A_23_P145397 | -0.1899 | 0.6398 | |
| SRP007169 | CCNC | 892 | RNAseq | -1.0241 | 0.0072 | |
| SRP008496 | CCNC | 892 | RNAseq | -1.0797 | 0.0000 | |
| SRP064894 | CCNC | 892 | RNAseq | -0.8034 | 0.0001 | |
| SRP133303 | CCNC | 892 | RNAseq | -0.2303 | 0.2364 | |
| SRP159526 | CCNC | 892 | RNAseq | -0.4986 | 0.0176 | |
| SRP193095 | CCNC | 892 | RNAseq | -0.8894 | 0.0000 | |
| SRP219564 | CCNC | 892 | RNAseq | -1.0281 | 0.0190 | |
| TCGA | CCNC | 892 | RNAseq | -0.0693 | 0.1519 |
Upregulated datasets: 0; Downregulated datasets: 3.
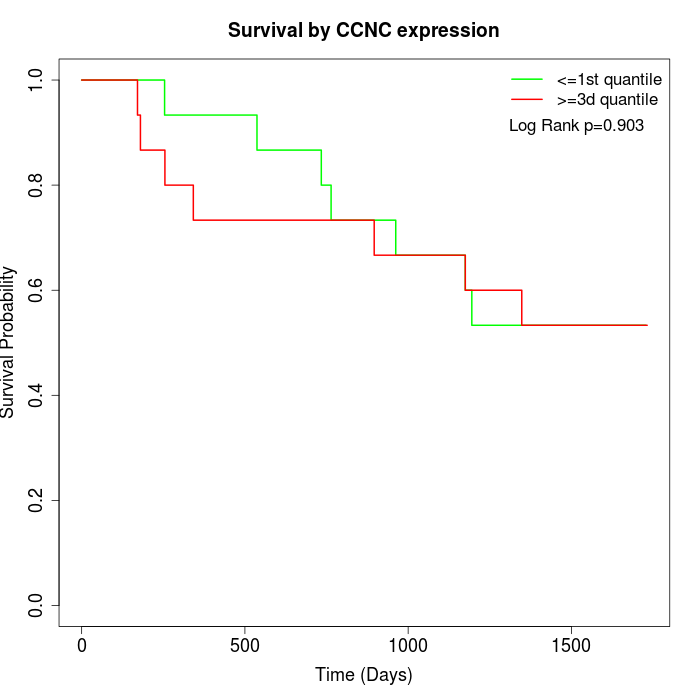
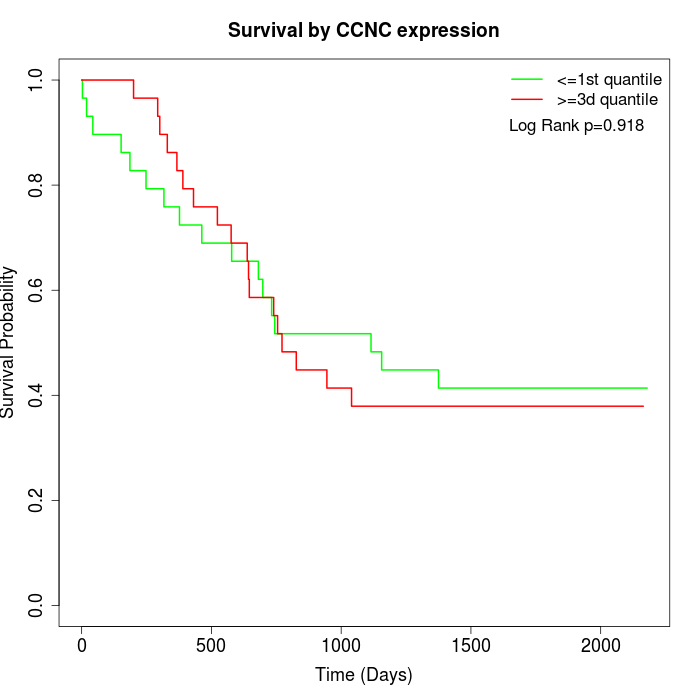
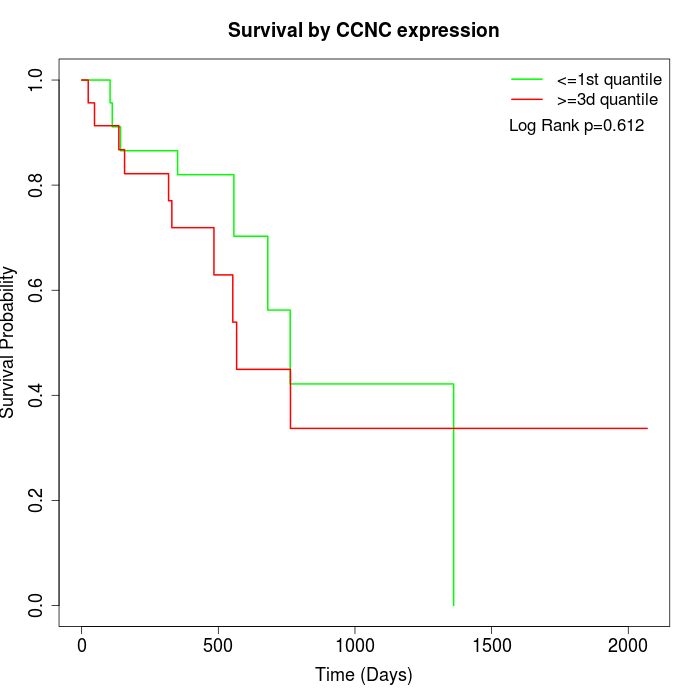
Survival by CCNC expression:
Note: Click image to view full size file.
Copy number change of CCNC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CCNC | 892 | 2 | 3 | 25 | |
| GSE20123 | CCNC | 892 | 2 | 2 | 26 | |
| GSE43470 | CCNC | 892 | 3 | 0 | 40 | |
| GSE46452 | CCNC | 892 | 2 | 11 | 46 | |
| GSE47630 | CCNC | 892 | 8 | 6 | 26 | |
| GSE54993 | CCNC | 892 | 3 | 2 | 65 | |
| GSE54994 | CCNC | 892 | 8 | 8 | 37 | |
| GSE60625 | CCNC | 892 | 0 | 1 | 10 | |
| GSE74703 | CCNC | 892 | 2 | 0 | 34 | |
| GSE74704 | CCNC | 892 | 1 | 0 | 19 | |
| TCGA | CCNC | 892 | 7 | 24 | 65 |
Total number of gains: 38; Total number of losses: 57; Total Number of normals: 393.
Somatic mutations of CCNC:
Generating mutation plots.
Highly correlated genes for CCNC:
Showing top 20/699 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CCNC | DNAJC7 | 0.784141 | 3 | 0 | 3 |
| CCNC | NPAT | 0.751892 | 3 | 0 | 3 |
| CCNC | ZNF638 | 0.750928 | 3 | 0 | 3 |
| CCNC | EIF5 | 0.740229 | 3 | 0 | 3 |
| CCNC | FAS | 0.737852 | 3 | 0 | 3 |
| CCNC | FUCA1 | 0.735936 | 3 | 0 | 3 |
| CCNC | ATG12 | 0.725132 | 4 | 0 | 3 |
| CCNC | EVC2 | 0.724561 | 3 | 0 | 3 |
| CCNC | MPI | 0.723099 | 3 | 0 | 3 |
| CCNC | DNAJB7 | 0.722781 | 3 | 0 | 3 |
| CCNC | NFKB1 | 0.722188 | 3 | 0 | 3 |
| CCNC | CRIM1 | 0.714855 | 3 | 0 | 3 |
| CCNC | FYTTD1 | 0.709905 | 4 | 0 | 3 |
| CCNC | CYP4F22 | 0.709476 | 3 | 0 | 3 |
| CCNC | DNAAF2 | 0.704745 | 3 | 0 | 3 |
| CCNC | GIN1 | 0.697737 | 4 | 0 | 4 |
| CCNC | MUC15 | 0.696588 | 3 | 0 | 3 |
| CCNC | ST13 | 0.689203 | 4 | 0 | 4 |
| CCNC | C6orf120 | 0.686767 | 7 | 0 | 6 |
| CCNC | HDAC3 | 0.685904 | 3 | 0 | 3 |
For details and further investigation, click here